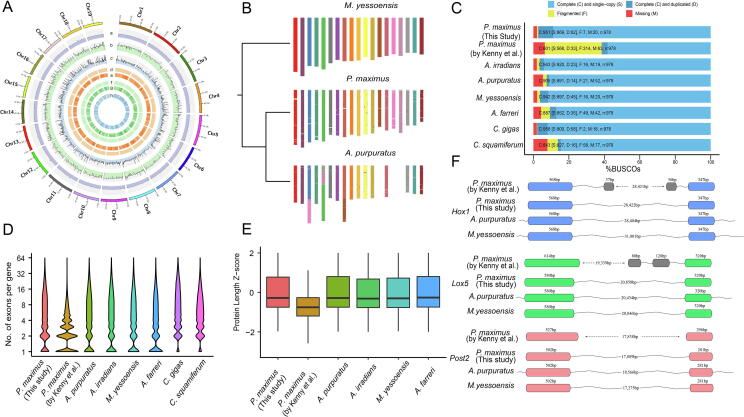
Fig. 1.
A: Global genome landscape of P. maximus. From outer to inner circles: GC content (a), depth of coverage of Illumina reads (b), depth of coverage of PacBio reads (c), distribution of homozygous (d) and heterozygous SNPs (e), distribution of homozygous (f) and heterozygous INDELs (g), distribution of genes (h). B: Macro-synteny of the P. maximus and A. purpuratus chromosomes to contemporary chromosomes of M. yessoensis. The conserved syntenic blocks are shown by the local fraction of genes from each M. yessoensis chromosomes. C: BUSCO evaluation on the predicted gene models. D: Distribution of number of exons per predicted gene model. E: Deviations of protein lengths for the single-copy orthologues in four scallops. F: Annotation of Hox1, Lox5, and Post2 gene structures in four scallop genomes. Exons from the same gene were annotated as distinct gene models in Kenny et al. (2020). Bins in grey indicate mis-annotated exons.