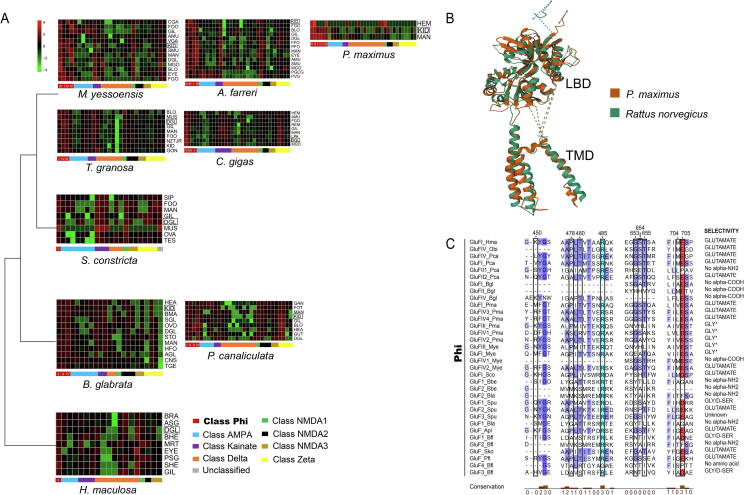
Fig. 3.
A: Expression patterns of iGluR genes among different tissues in P. maximus and eight molluscs. B: Protein structure alignment of P. maximus GluFIII and R. norvegicus GluK2 (7KS3). C: Multiple protein alignment of Phi class of iGluR gene residues involved in ligand-binding. Numbers shown on top correspond to GluA2 of Rattus norvegicus (P19491). Residues involved in ligand binding are indicated by a black frame. Acid and basic amino acid residues were highlighted by red and light blue, respectively. The levels of conservation are denoted by blue background and a bar chart at the bottom. Agonists predicted for iGluRs with non-conservative residues at 653 and 655 were indicated with an asterisk. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)