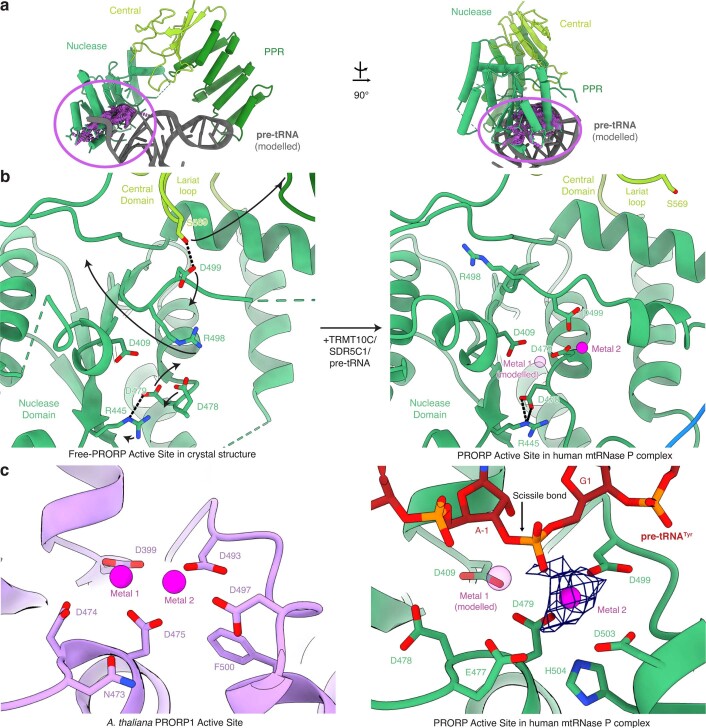
Extended Data Fig. 7. Comparison of inactive and active conformations of PRORP.
a, Clashes between metallonuclease domain of free PRORP in the closed conformation and the substrate pre-tRNA. The crystal structure of free PRORP (PDB: 4XGL)23 was superimposed via its PPR domain to PRORP in mtRNase P complex. Pre-tRNATyr, shown in gray, was modeled based on its binding site in mtRNase P complex. Clashes between pre-tRNA and free PRORP are indicated in purple. b, Comparison of the PRORP nuclease active site observed in the crystal structure of free PRORP (PDB: 4XGL)23 and in the active conformation in the mtRNase P complex. Arrows indicate the rearrangements that may occur in the free PRORP structure upon binding to the TRMT10C-SDR5C1 complex and substrate RNA. Active site aspartates are shown as sticks. Metal 1 was modeled based on Mn2+ binding sites in the At-PRORP1 crystal structure (PDB: 4G24)41. The pre-tRNA is omitted for clarity. c, Comparison of the active sites of At-PRORP1 (PDB: 4G24)41 (left) and active PRORP (right). The two metal ions in the At-PRORP1 structure are shown as spheres. The cryo-EM density map for metal 2 in the PRORP active site is shown as blue mesh.