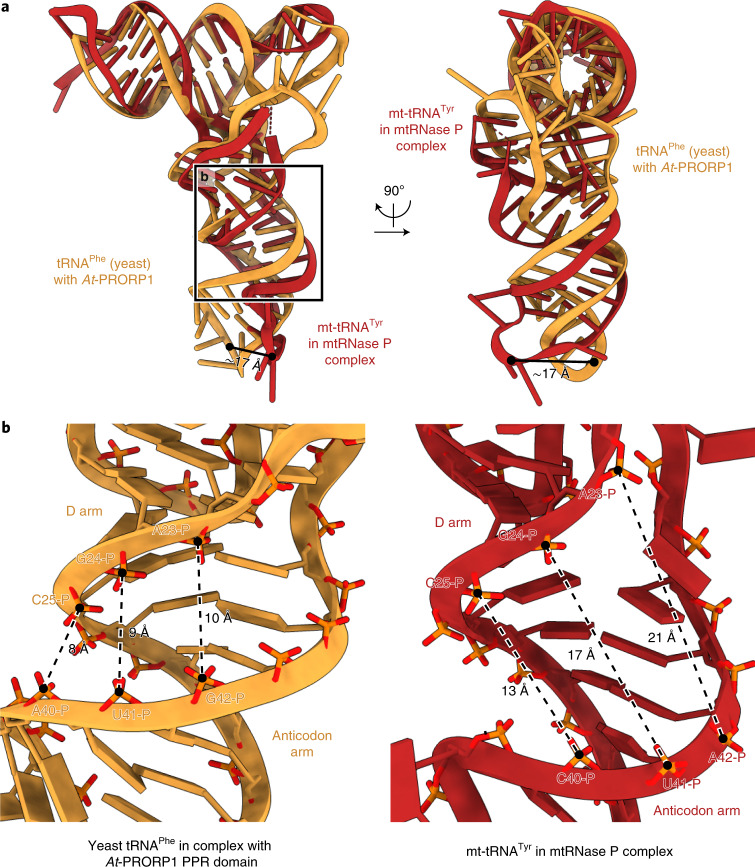
Fig. 3. The pre-tRNA adopts a distorted structure.
a, Comparison of tRNA structures for human pre-(mt)tRNATyr in the mtRNase P complex (red) and yeast tRNAPhe in complex with the A. thaliana PRORP1 (At-PRORP1) PPR domain (orange) (PDB 6LVR)27. The two tRNA structures are aligned with the acceptor and T arms (pre-tRNATyr: residues 1–7 and 44–65; tRNAPhe: residues 1–7 and 49–72). In the mtRNase P complex, the pre-tRNATyr adopts a distorted structure compared to a canonical tRNA, resulting in an approximately 17 Å displacement of the anticodon loop (distance between phosphate atoms between anticodon residues 1 and 2, shown as black circles). b, Distance comparison between the D arm and the anticodon arms of pre-tRNATyr in the mtRNase P complex and tRNAPhe bound to the At-PRORP1 PPR domain. Distances between phosphorous atoms of nucleotide pairs 25–40, 24–41 and 23–42 (canonical tRNA numbering33) in both tRNAs are shown as dashed lines. The distance between the D and anticodon arms of pre-tRNATyr in the mtRNase P complex (right) is considerably larger than in a tRNA bound to At-PRORP1 (left).