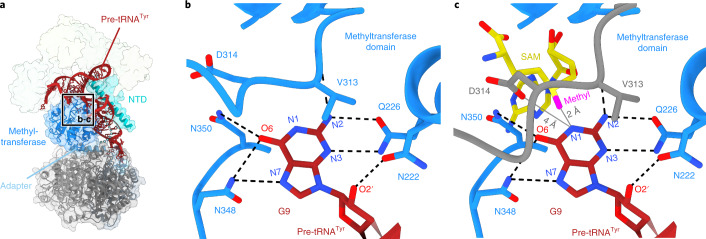
Fig. 4. The TRMT10C methyltransferase active site.
a, Overview of the TRMT10C–SDR5C1 subcomplex with the region enlarged in b and c indicated. TRMT10C, SDR5C1 and pre-tRNATyr are shown as ribbons, with TRMT10C, SDR5C1 and PRORP additionally shown as transparent surfaces. b, Close-up view of the methyltransferase active site of TRMT10C. Nucleotide G9 of the pre-tRNATyr and active site residues within 4 Å of G9 and D314 of TRMT10C are shown in stick representation. Potential hydrogen bonds are shown as dashed black lines. Methyl-acceptor atom N1 and additional atoms of G9 potentially involved in active site interactions are labeled. c, Precatalytic model of the TRMT10C methyltransferase active site for G9 methylation. To generate this model, the crystal structure of the substrate-free SAM-bound TRMT10C methyltransferase domain (PDB 5NFJ)19 was superimposed with the TRMT10C methyltransferase domain in mtRNase P. The model is composed of the active site of substrate-bound TRMT10C in the mtRNase P complex, shown as in b, with the location of the SAM cofactor and the conformation of residues 310 to 320 (motif II loop) of TRMT10C modeled as in the SAM-bound crystal structure. SAM is shown in yellow with the methyl group in magenta, and the motif II loop derived from PDB 5NFJ is shown in gray.