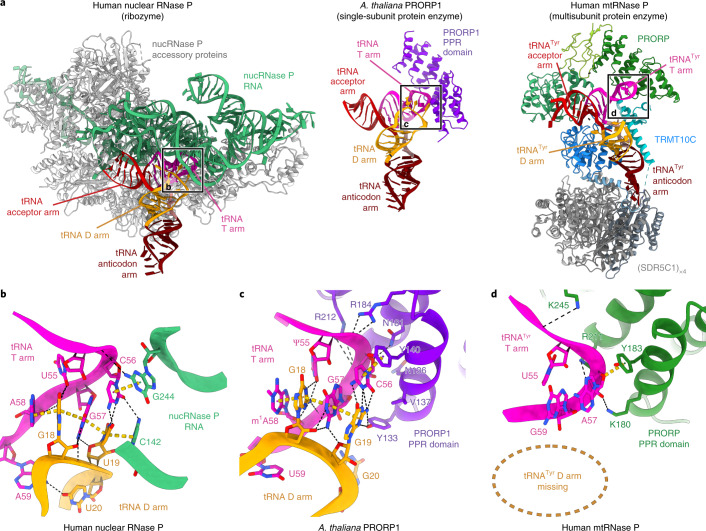
Fig. 6. Comparison of substrate recognition by different RNase P enzymes.
a, Structural comparison between human nuclear RNase P ribozyme (PDB 6AHU)26, Arabidopsis thaliana PRORP1 PPR domain in complex with tRNA (PDB 6LVR)27 and human mtRNAse P in complex with pre-tRNATyr (left to right). Nuclear RNase P (nucRNase P) catalytic RNA and accessory proteins are colored in green and gray, respectively; At-PRORP1 PPR domain is colored in purple; and mtRNase P protein domains are colored as in Fig. 1a. The tRNAs are shown colored by structural elements: acceptor arm, red; T arm, pink; D arm, yellow; anticodon loop, dark red; variable region, light brown (not labeled). Elbow regions of tRNAs, enlarged in panels b–d, are indicated. b, Close-up of the tRNA-elbow recognition site in human nuclear RNase P. Bases in the elbow region of the tRNA and RNase P RNA bases within 4 Å are shown as sticks. The conserved interaction between the D and T arm is recognized by the RNA component of RNase P. c, Close-up of the tRNA-elbow recognition site in At-PRORP1. Bases in the elbow region of tRNA and At-PRORP1 residues within 4 Å are shown as sticks. The D and T arm interaction, including the C56-G19 base pair contact, is recognized by the PPR domain of At-PRORP1. d, Close-up of the tRNA-elbow binding site in the human mtRNase P complex. Bases in the elbow region of pre-tRNATyr and PRORP residues within 4 Å are shown as sticks. Interactions between T and D arms in the elbow region are absent, and PRORP PPR domain residues interact primarily with the RNA backbone.