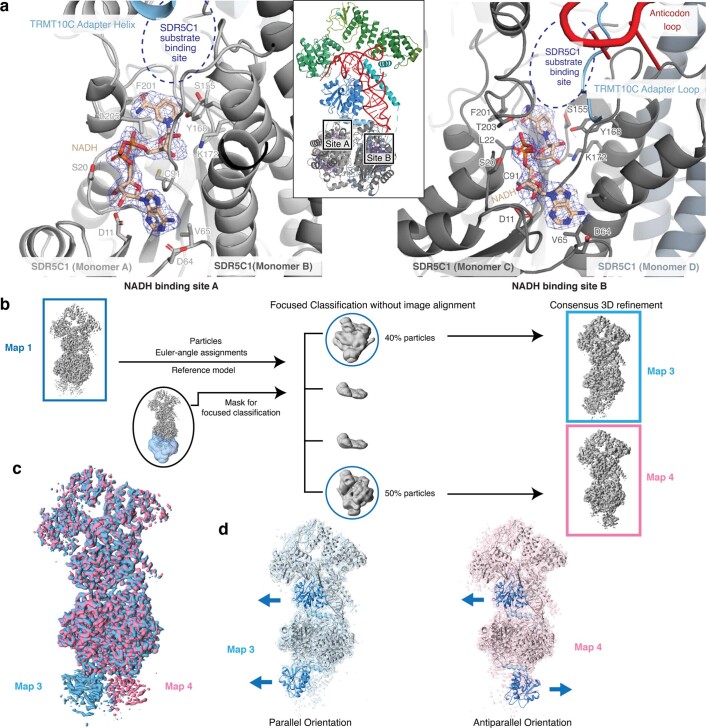
Extended Data Fig. 3. Structural details of the TRMT10C-SDR5C1 subcomplex.
a, NADH bound to the two non-equivalent NADH binding sites in the SDR5C1 tetramer. NADH and residues within 3.5 Å are shown as sticks. The cryo-EM density around NADH is shown as blue mesh. The NADH binding site A (left) is located near the C-terminal end of the TRMT10C adapter helix, while the NADH binding side B (right) is located near the TRMT10C adapter loop and anticodon recognition site. The dehydrogenase substrate binding sites near both NADH binding sites are partially occluded by the adapter region of TRMT10C and/or tRNA. b, Cryo-EM processing workflow to resolve the two opposing orientations of the second TRMT10C monomer bound to SDR5C1 tetramer. c, Overlay of the two reconstructions map 3 and map 4, showing opposing orientations of the bottom density. Surface color of the maps correspond to the color used for boxes around map 3 and map 4 in (b). d, Models of mtRNase P with a second TRMT10C monomer (residues 175–385) bound to SDR5C1 tetramer in two opposing orientations fitted into cryo-EM density maps from (b) colored as in (c). Domain coloring of TRMT10C between residues 175 to 385 as in Fig. 1(d); models for all remaining regions are shown in gray. The adapter helix of TRMT10C can bind to the groove on SDR5C1 tetramer in two 180°-rotated orientations resulting in either parallel orientation or antiparallel orientation of TRMT10C protomers. Arrows next to each TRMT10C monomer denote the general orientation of the N and C termini, with the arrowhead denoting the C-terminus.