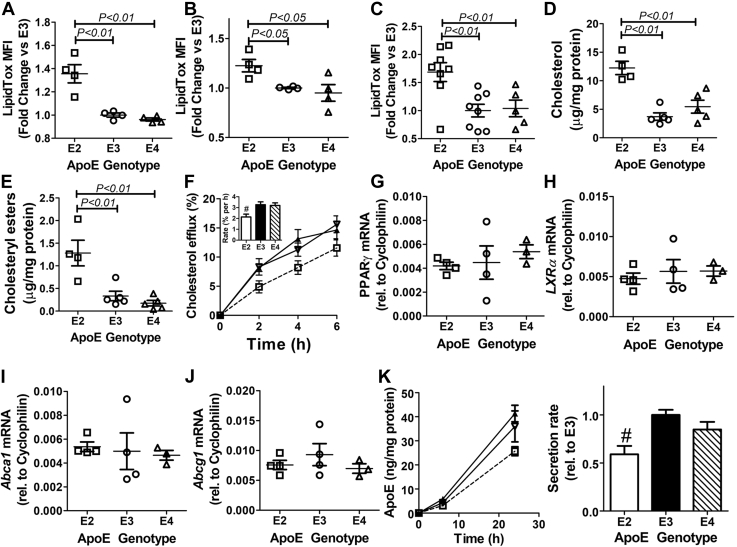
Figure 4.
Cholesterol enrichment in apoE2-expressing myeloid cells due to impairment of cholesterol efflux and apoE secretion. The fluorescent dye LipidTOX was used to assess neutral lipid accumulation in (A) peripheral blood monocytes (N = 4), (B) peritoneal macrophages (N = 4), and (C) lineage-negative bone marrow cells (N = 8) isolated from APOE2, APOE3, and APOE4 gene replacement mice. D, free cholesterol and (E) cholesteryl ester contents were measured in peritoneal macrophages isolated the gene replacement mice (N = 5) by biochemical methods. F, the peritoneal macrophages from APOE2 (□, open bar), APOE3 (▲, filled bar), and APOE4 (Δ, hatched bar) were preloaded with [3H]cholesterol followed by incubation with HDL to assess cholesterol efflux. The data represent the mean ± SD using four mice per group. The inset shows the rate of cholesterol efflux calculated by linear regression. # indicates p < 0.05 difference from the other groups. mRNA was also isolated from macrophages of the gene replacement mice (N = 4) for amplification of PPARγ (G), LXRα (H), ABCA1 (I), and ABCG1 (J) mRNA. Expression levels were normalized to the cyclophilin mRNA levels in each sample. K, media were collected from cultured peritoneal macrophages from APOE2 (□), APOE3 (▲), and APOE4 (Δ) mice (N = 6) for apoE quantification by ELISA. The graph on the right shows the rate of apoE secretion relative to the rate observed with apoE3-expressing cells. # indicates p < 0.05 difference from the other groups. All data were evaluated by one-way ANOVA with Student–Newman–Keuls post hoc test for significant differences as indicated. ABCA1, ATP-binding cassette-A1; ABCG1, ATP-binding cassette-G1; apoE, apolipoprotein E; HDL, high-density lipoprotein; LXRα, liver X receptor-α; PPARγ, peroxisome proliferator–activated receptor-γ.