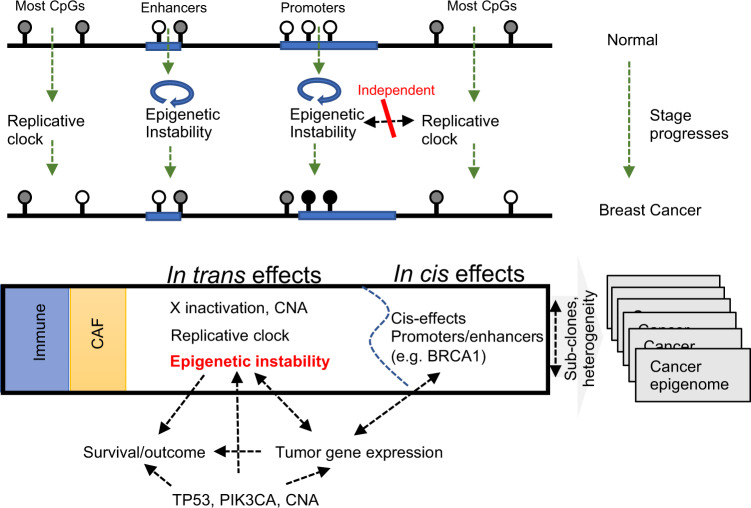
Fig. 5. Unified model delineating the multi-factorial processes giving rise to breast cancer DNA methylation.
As the carcinogenesis process progresses (top), epigenomes are affected by replication-dependent methylation loss in most of the genome, and by a second, uncorrelated epigenetic instability process modulating methylation in promoters and enhancers. When cancer epigenomes are surveyed (middle), the observed profiles involve a superposition of TME signatures, with the patient-specific replication and instability signatures, and with epigenetic dosage compensation. These processes are each affecting a large number of genomic loci through one common mechanism (in trans effects). Additional localized patient-specific methylation aberrations are uncorrelated with these in trans effects and may regulate gene expression in cis. Deconvolution of these multi-layered effects shows linkage between epigenetic instability and disease stage and prognosis (bottom). Cancer epigenomic heterogeneity is also induced by cellular heterogeneity (such as clonal structure).