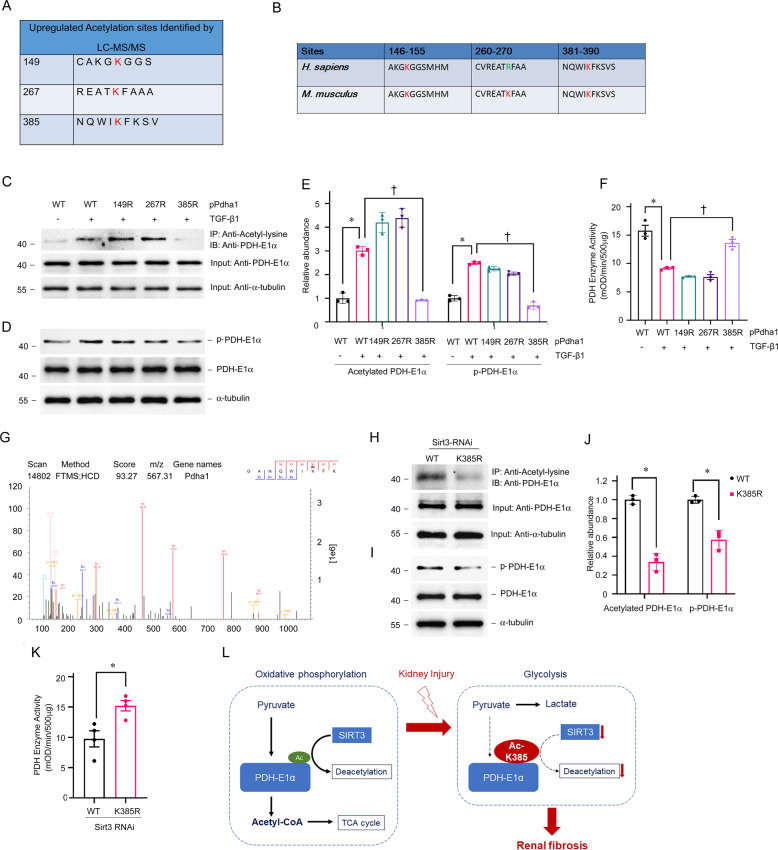
Fig. 8. Discovery of acetylated sites of PDHE1α as a target of SIRT3 in TECs.
A Summary of peptide fragments of upregulated acetylated lysine residues of PDHE1α; B Alignment of sequences around K149, K267, K385 of human and mouse PDHE1α; C–E TECs transfected with expression vectors for WT PDHE1α, PDHE1α single acetylation-point mutant K149R, K267R, or K385R, then stimulated with TGF-β1. Levels of acetylated PDHE1α (C, E), phosphorylated PDHE1α (D, E), and PDH enzyme activity (F) in cell lysates. PDHE1α served as the standard (*P < 0.05, †P < 0.05; n = 3). G Representative m/z spectra (K385) determined from mass spectrometry analyses of PDHE1α. H–K TECs transfected with Sirt3 siRNA followed by expression vectors for WT PDHE1α or PDHE1α K385R. Mitochondrial lysates immunoprecipitated with anti-acetyl-lysine antibody and analyzed with anti-PDHE1α. PDHE1α served as the standard (*P < 0.05; n = 3) (H, J); Western blots of phosphorylated PDHE1α and total PDHE1α in TECs in Sirt3 knockdown TECs transfected with WT or PDHE1α K385R. PDHE1α served as the standard (*P < 0.05; n = 3) (I, J). K Enzyme activity of PDH in Sirt3 knockdown TECs with PDHE1α K385R transfection vs. WT (*P < 0.05 vs. WT transfection, n = 4). L Diagram depicts the role of SIRT3 in the control of PDHE1α deacetylation in renal fibrosis.