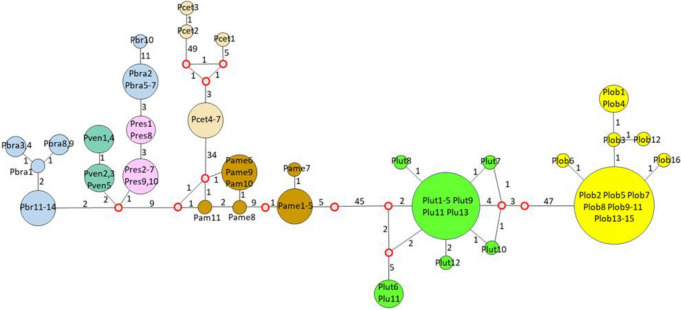
Figure 3.
Neighbor-joining analysis of the Gp43 partial DNA sequences revealed several haplotypes. The size of the spheres is proportional to the number of individuals in each circle (numbers are shown inside the spheres). Median vectors (red open rings) represent missing or extinct species. Numbers between haplotypes indicate mutational steps. Large number of mutations were observed between Paracoccidioides lutzii (Plut-green) and P. loboi (Plob-yellow) (n = 54), between these 2 haplotypes and P. americana (Pame-brown) and P. cetii (Pcet-beige) (n = 52). Thirty-four mutations separate P. americana from P. cetii. In contrast, few mutations were observed between these four haplotypes and the remaining Paracoccidioides species (Pbra = P. brasiliensis [blue], Pres = P. restrepiensis [pink], Pven = P. venzuelensis [green]) (n = 11). Three Japanese dolphin haplotypes (Pcet = P. cetii [beige 1 to 3]) showed three missing or extinct haplotypes between them and the four USA dolphins (Pcet = P. cetii 4–7-beige) in this study.