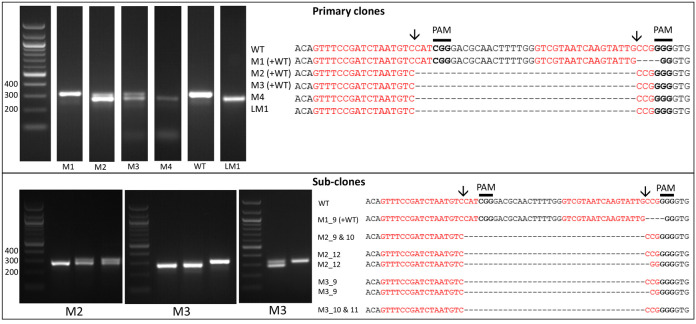
Figure 2. Screening of the CRISPR-Cas induced genome editing in T. pseudonana.
Screening by PCR and sequencing. Expected sgRNA cut indicated by ↓. Red text shows the sgRNA target sequence and bold text the PAM motif. Primary clones: Several primary clones contain sequences showing both CRISPR-induced mutations and the wildtype (WT) sequence, as seen by the presence of two bands following PCR, these are indicated by (+WT). Remaining samples represent different cell lines. M1 shows a 4 nt deletion from the second sgRNA whilst mutants M2-M4 show a 37 nt deletion between the two CRISPR-Cas cut sites. Sub-clones: The gel shows examples of a selection of sub-clones derived from the primary clones. Sub-clones are labelled according to the primary clone and sub-clone number. With the exception of M1_9, which gives a WT sequence and 4 nt deletion as seen in the primary clone, all sub-clones chosen for sequencing are bi-allelic. Two-thirds of sequenced bi-allelic sub-clones show a single sequence with a 37 nt deletion suggesting that both alleles carry the same mutation. In sub-clones where mutations differ between alleles both sequences are shown.