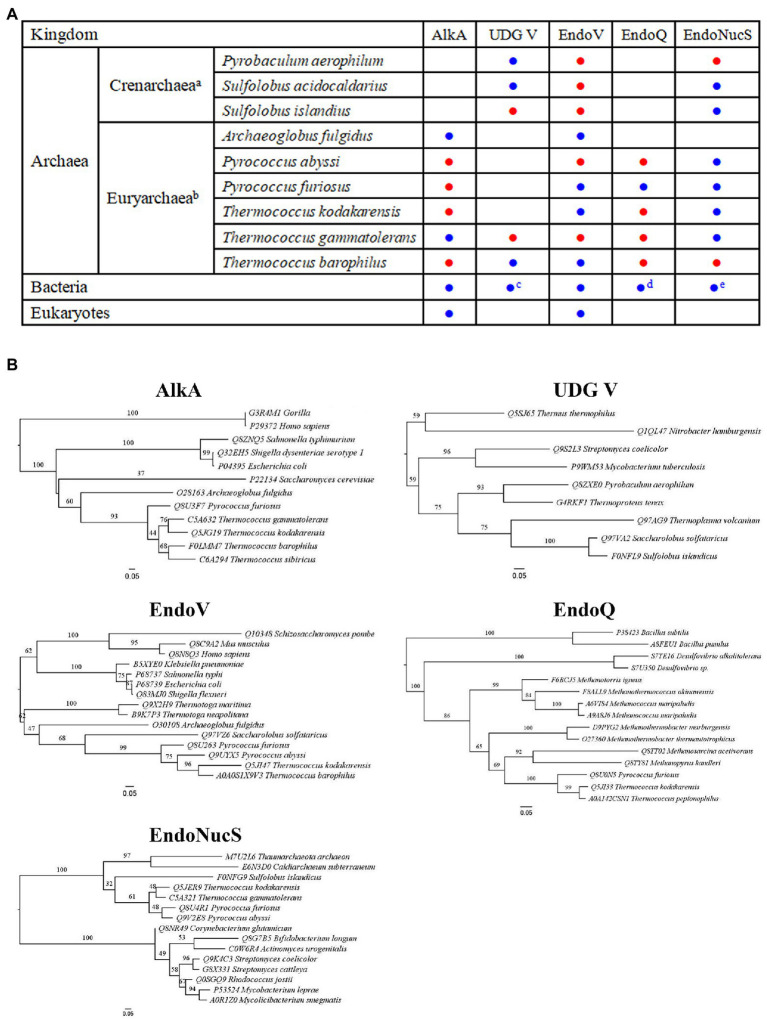
Figure 1.
Distribution and phylogenetic analyses of DNA repair proteins involved in Hypoxanthine (Hx) repair. (A) Distribution of 3-methyladenine DNA glycosylase II (AlkA), uracil DNA glycosylase (UDG) V, Endonuclease V (EndoV), Endonuclease Q (EndoQ), and Endonuclease NucS (EndoNucS). a and b: only includes archaea which at least one enzyme among five target enzymes has been characterized; c: includes a few hyperthermophilic bacteria; d: includes partial bacteria, such as Bacillus and Desulfovibrio; and e: includes partial bacteria that lack mismatch repair pathway, such as Actinobacteria. The characterized and uncharacterized enzymes are shown with blue and red circle, respectively. (B) Phylogenetic analyses of AlkA, UDG V, EndoV, EndoQ, and EndoNucS from hyperthermophilic Archaea. Note that only partial Archaea, bacteria, and eukaryotes are included in these five phylogenetic trees due to the space limit.