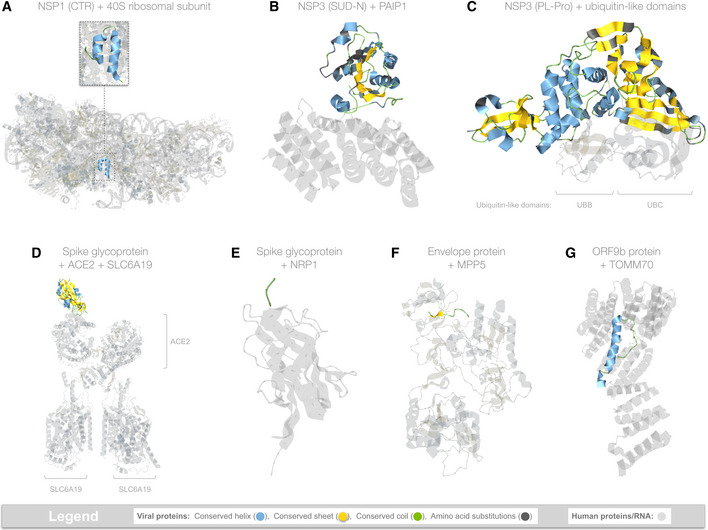
Figure 3. Viral hijacking of human proteins.
-
AHijacking of ribosomal complexes is shown in 14 matching structures, most of which were determined using the full‐length sequence of NSP1 (180 residues); however, only a ˜36 residue fragment was ordered enough to appear in the structures. The coloring scheme highlights the location of this fragment within the ribosome (https://aquaria.ws/P0DTC1/6zlw), revealing how NSP1 blocks host mRNA translation (Thoms et al, 2020).
-
BHijacking of PAIP1 (a.k.a. “PABP‐interacting protein 1”) is shown in only one matching structure that was determined using the SUD‐N region of NSP3 from SARS‐CoV (Nikulin et al, 2021). The structure (https://aquaria.ws/P0DTC1/6yxj) shows the strong overall sequence similarity in SARS‐CoV‐2 and reveals that, of the 15 residues contacting PAIP1, 13 are identical in SARS‐CoV‐2.
-
CHijacking of ubiquitin‐like (Ubl) domains is shown in 10 matching structures, of which only one showed simultaneous binding to two Ubl domains (shown above). The structure (https://aquaria.ws/P0DTC1/5e6j) was determined using NSP3 from SARS‐CoV (Békés et al, 2016), which had strong overall sequence similarity in SARS‐CoV‐2; of the 31 residues contacting UBB or UBC, 27 are identical in SARS‐COV‐2.
-
DHijacking of ACE2 is shown in 46 matching structures; however, only two also show binding to SLC6A19 (Yan et al, 2020). In the structure shown here (https://aquaria.ws/P0DTC2/6m17), spike glycoprotein does not directly bind to SLC6A19.
-
EHijacking of NRP1 (a.k.a. neuropilin‐1) is shown in only one matching structure (https://aquaria.ws/P0DTC2/7jjc), which includes only a three‐residue region from spike glycoprotein (Daly et al, 2020).
-
FHijacking of MPP5 (a.k.a. PALS1, “protein associated with Lin‐7 1”) is shown in only one matching structure (https://aquaria.ws/P0DTC4/7m4r), which includes only a nine‐residue region from envelope protein (Liu & Chai, 2021).
-
GHijacking of TOMM70 (a.k.a. “translocase of outer mitochondrial membrane protein 70”) is shown in only one matching structure (https://aquaria.ws/P0DTD2/7kdt), which includes only a 38‐residue region from ORF9b protein (Gordon et al, 2020).
Data information: Made using Aquaria and Keynote.
