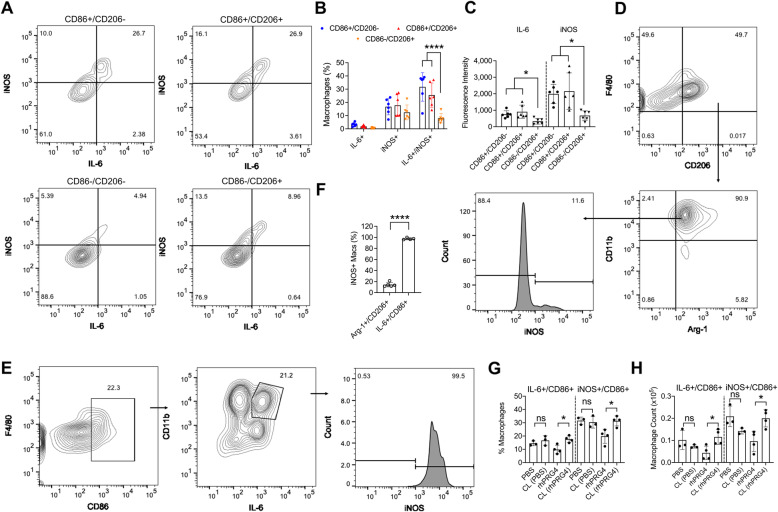
Fig. 2.
Characterization of the expression of inflammatory markers, interleukin-6 (IL-6), and inducible nitric oxide synthase (iNOS) in gated synovial macrophages that express CD86 and/or CD206 surface markers from 2-month-old Prg4 gene-trap (Prg4GT/GT) animals and impact of intra-articular (IA) recombinant human proteoglycan-4 (rhPRG4) treatment on IL-6 and iNOS expression in CD86+ macrophages. Macrophages were identified as CD11b+ F4/80+ MHC class II− CD86+ and/or CD206+ and the expression levels of IL-6 and iNOS (both are markers of inflammatory M1 macrophages) and arginase-1 (Arg-1) (a marker of anti-inflammatory M2 macrophages) were evaluated using intracellular staining with PE-anti-IL-6, Alexa Fluor 488-anti-iNOS, and PE-anti-Arg-1 antibodies. Positivity thresholds were set using fluorescence minus one (FMO) panel controls. rhPRG4 (1 mg/mL; 10μL) or phosphate-buffered saline (PBS) (10μL) were administered IA in 2-month-old Prg4GT/GT animals and analysis of IL-6 and iNOS positivity in CD11b+ F4/80+ MHC class II− CD86+ macrophages was performed at 24 h following IA treatments. Macrophage cell counts were determined using precision counting beads and expressed as numbers per joint. Statistical analyses of IL-6 and iNOS staining intensities in macrophages and percentages of IL-6+ and/or iNOS+ macrophages were performed using one-way and two-way ANOVA, respectively. Tukey’s post hoc test was used in one- and two-way ANOVAs. Analysis of iNOS positivity in Arg-1+/CD206+ and IL-6+/CD86+ macrophages and impact of rhPRG4 treatment was performed using unpaired and paired Student’s t test, respectively. Experimental groups included three (2 males and one female) to six animals (3 males and 3 females). In all experiments, independent biological samples were analyzed in duplicates, and the averages of both technical replicates were included in the analysis. ns non-significant; *p<0.05 and ****p<0.0001. A A representative flow cytometry contour plot of IL-6 and iNOS probing in CD86+ and/or CD206+ macrophages. Gating for CD86+ and/or CD206+ macrophages was performed according to supplementary figure 2 and fig. 1B and cells in each quadrant were plotted according to their IL-6 and iNOS staining intensities, and percentages of IL-6+, iNOS+, and IL-6+/iNOS+ macrophages were determined. B Percentages of dually positive IL-6 and iNOS CD86+/CD206− and CD86+/CD206+ macrophages were higher than dually positive IL-6 and iNOS CD86−/CD206+ macrophages. C Mean IL-6 and iNOS staining intensities were higher in CD86+/CD206− and CD86+/CD206+ macrophages compared to CD86−/CD206+ macrophages. D Gating strategy to identify Arg-1+/CD206+ macrophages in synovial tissues from 2-month-old Prg4GT/GT joints. Singlets and viable cells were identified as shown in Supplementary Figure 2. Viable cells were gated according to their CD206 and F4/80 expression status. Dually positive cells were then gated for Arg-1 and CD11b to identify Arg-1+/CD206+ macrophages. The percentage of the iNOS+ subset was subsequently determined. E Gating strategy to identify IL-6+/CD86+ macrophages in synovial tissues from 2-month-old Prg4GT/GT joints. Singlets and viable cells were identified as shown in Supplementary Figure 2. Viable cells were gated according to their CD86 and F4/80 expression status. Dually positive cells were then gated for IL-6 and CD11b to identify IL-6+/CD86+ macrophages. The percentage of the iNOS+ subset was subsequently determined. F The percentage of iNOS+/IL-6+/CD86+ macrophages was higher than the corresponding percentage in iNOS+/Arg-1+/CD206+ macrophages. G rhPRG4 treatment reduced the percentages of IL-6+/CD86+ and iNOS+/CD86+ macrophages compared to contralateral (CL) joints. No reductions were observed with PBS treatment. H rhPRG4 treatment reduced the numbers of IL-6+/CD86+ and iNOS+/CD86+ macrophages compared to contralateral (CL) joints. No reductions were observed with PBS treatment