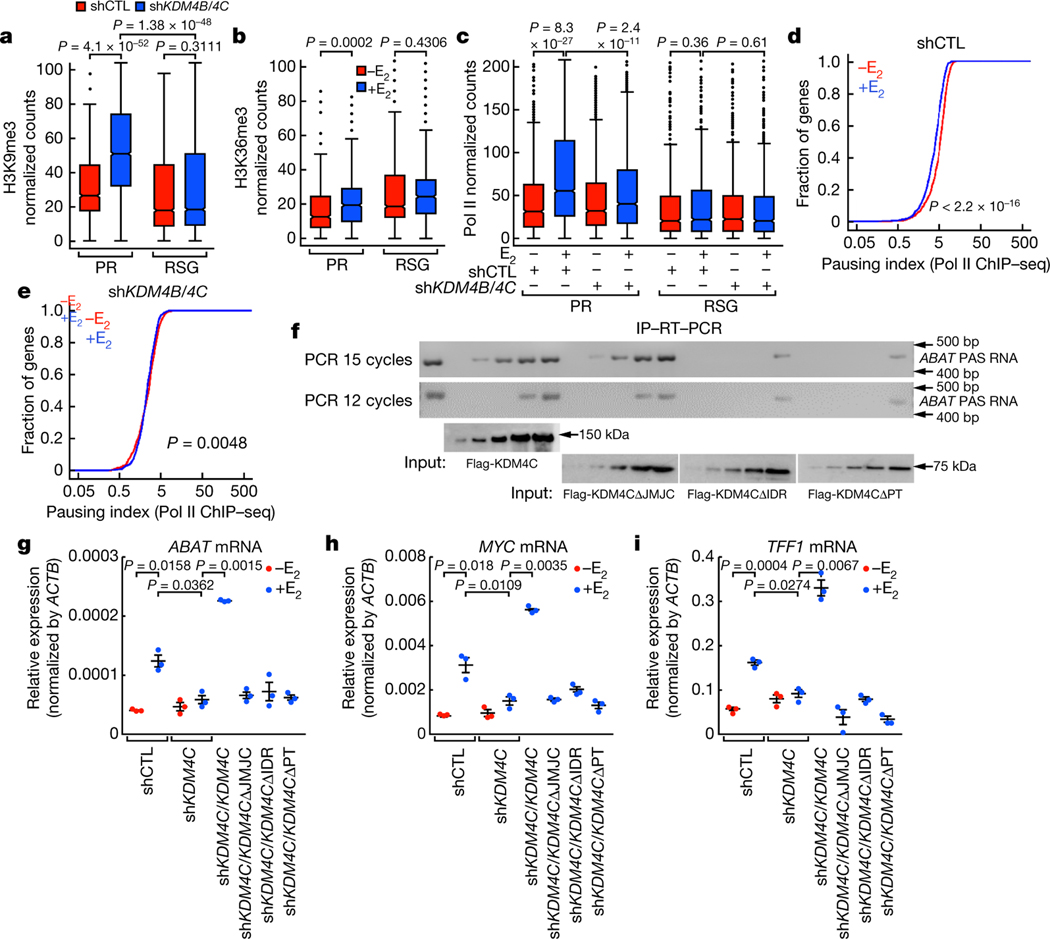
Fig. 4 |. PAS RNA stabilization of KDM4B and KDM4C at the promoter.
a-c, Box plot analysis representing the effect of KDM4B/KDM4C knockdown on the accumulation of H3K9me3 (a), the effect of E2 on H3K36me3 (b) at the indicated promoters, and the effect of KDM4B/KDM4C knockdown on Pol II occupancy over the gene bodies of the indicated genes (c). The box plot parameters are the same as in Fig. 1 (n = 2 independent experiments). The P values were determined by two-sided Wilcoxon test. d, e, Cumulative distribution of the Pol II pausing ratio based on Pol II ChIP-seq analysis (n = 2 independent experiments) of PR genes following E2 treatment in short hairpin control (shCTL) MCF-7 cells (d) or shKDM4B/KDM4C MCF-7 cells (e). The P values were determined by two-sided Kolmogorov-Smirnov test. f, In vitro immunoprecipitation (IP)-RT-PCR data showing the required intrinsically disordered region (IDR) of KDM4C for interaction with ABAT PAS RNA. The experiment was repeated three times with similar results. Gel source data are available in Supplementary Fig. 1. Flag-KDM4CΔPT denotes flag-tagged KDM4C-mutant protein lacking both the PHD and Tudor domain. g-i, RT-qPCR data showing the rescue effect of full-length KDM4C or the KDM4C mutants on the indicated gene expression (ABAT (g), MYC (h) and TFF1 (i)). Data are shown as individual values, mean ± s.d. (n = 3). The P values were determined by two-sided Student’s t-test.