Figure 1.
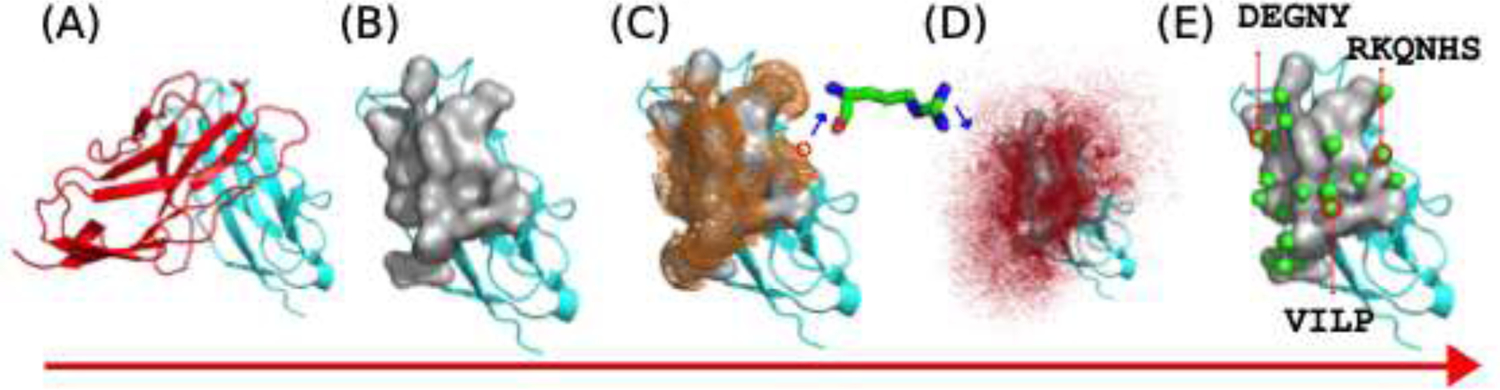
Reside specific pharmacophore generation using ProtLID. (A) identifying the protein binding interface. (B) Surface representation of the interface. (C) Mapping hypothetical starting points or reside probe sampling. (D) Residue binding preferences obtained from MD simulations (E) Predicted pharmacophore (complementary reside binding pattern, with locations (green spheres) and residue types (listed).
