Figure 2.
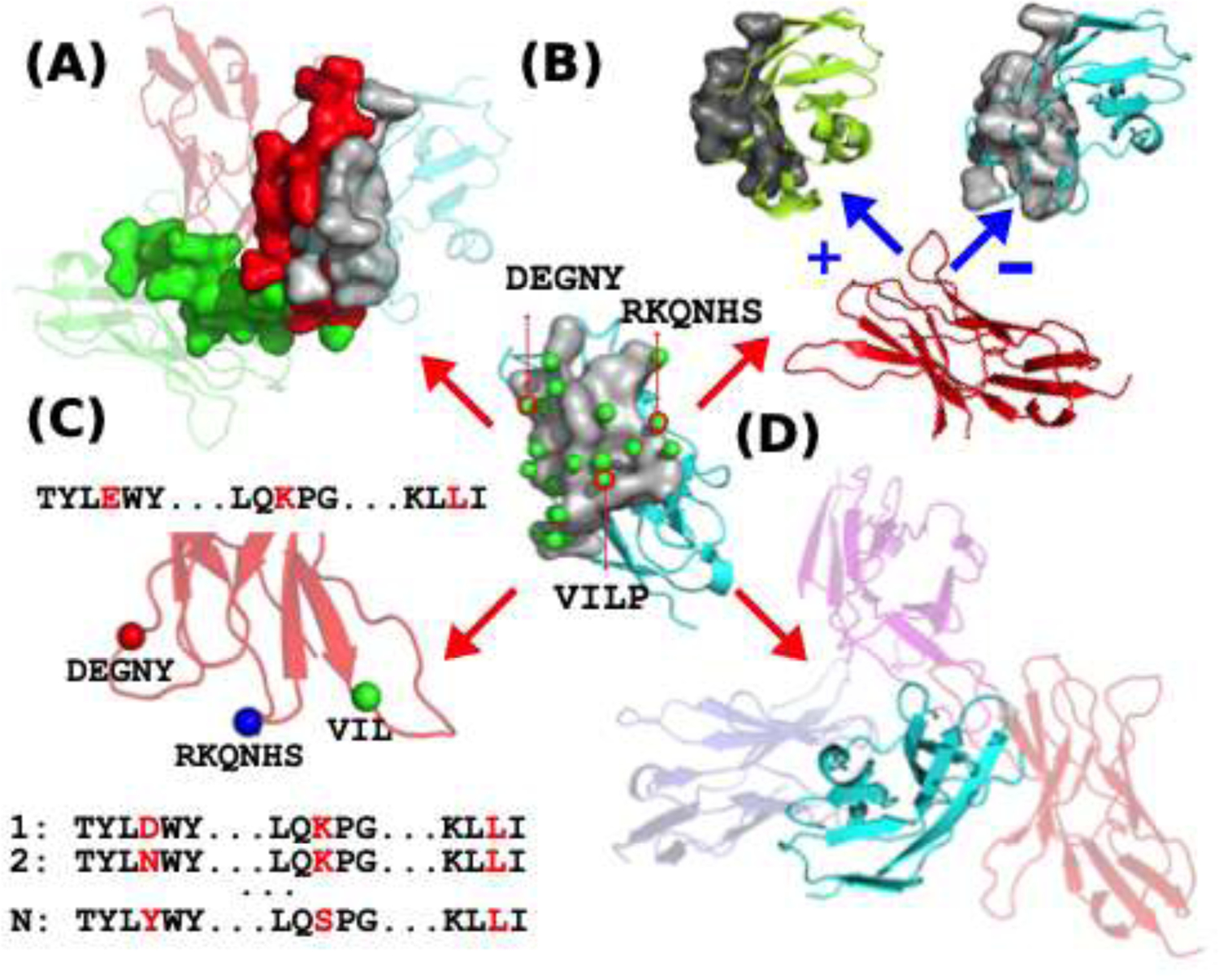
Possible uses of ProtLID in protein interface modeling. (A) Identifying cognate ligands for a receptor. Receptor (in cyan) interface is in grey. ProtLID predicts a residue based pharmacophore that fits (red interface) the best the cognate ligand among many candidates (such as green). (B) Redesigning protein interface for selectivity. Plus and minus signs refer to increased and decreased binding, as a function of better or poorer fit to the calculated pharmacophore. (C) Library design for phage display studies. Rs-pharmacophore directs the library construction (position and allowed mutations) for subsequent phage display studies (D) Docking two proteins. Randomly exploring possible binding interfaces for a cognate ligand.
