Figure 2.
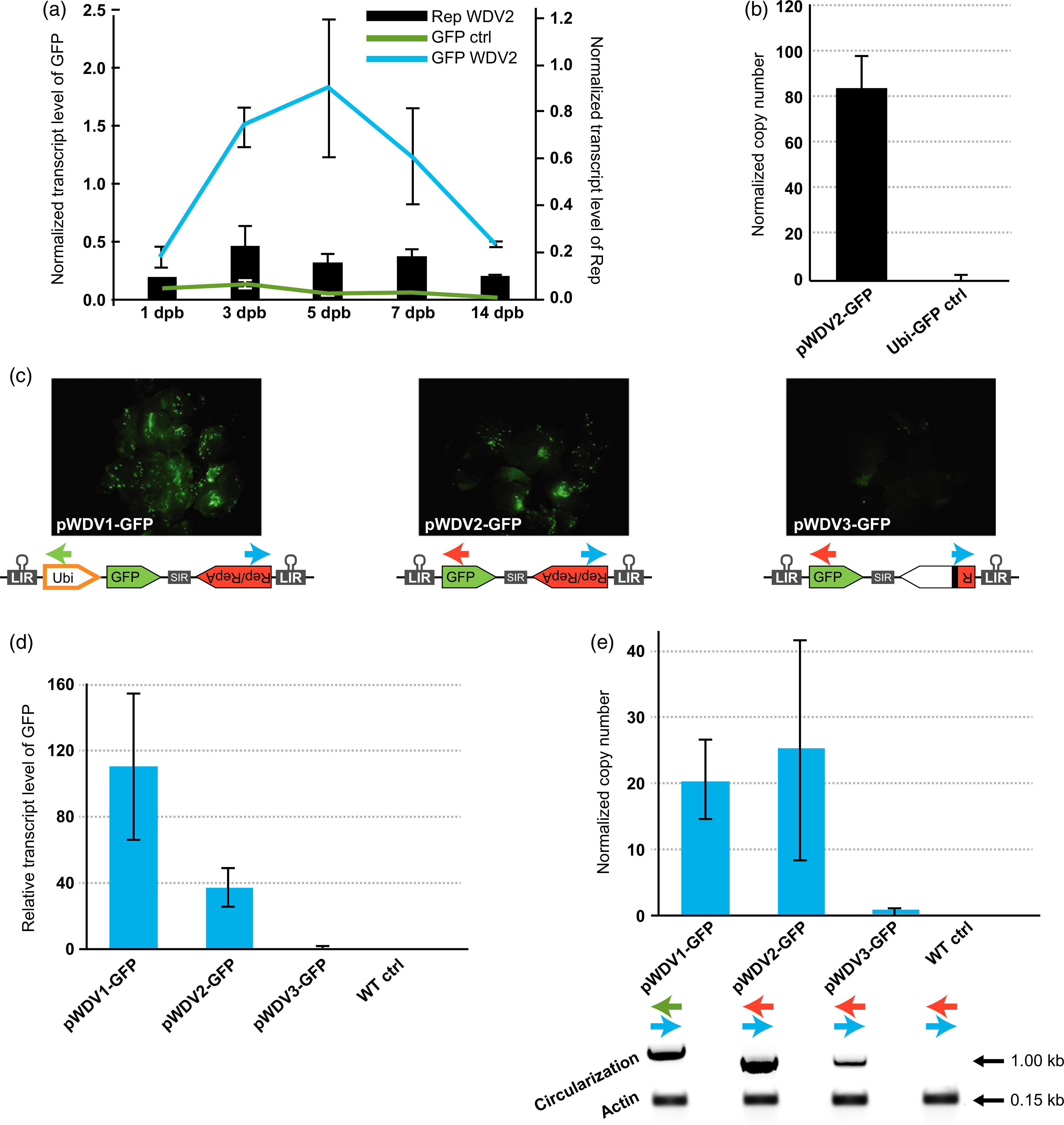
Wheat dwarf virus (WDV) replicon replication and performance of different WDV architectures designed for genome engineering of cereal species. (a) Time course of WDV replication and relative transcript level of GFP and Rep/RepA proteins in wheat calli at different days post bombardment (dpb). (b) Normalized copy number of the GFP-containing replicon relative to the Ubi-GFP control at 5 dpb. (c) GFP expression in wheat calli using the different WDV architectures: WDV1-GFP (left panel), WDV2-GFP (central panel) and WDV3-GFP (right panel). (d) qRT-PCR quantification of transcript level normalized to the non-replicating pWDV3-GFP. (e) Replicon copy number normalized to the non-replicating pWDV3-GFP and detection of circularization of the replicon and the actin PCR control. The colored arrows in (c) indicate the primers used to detect circularization in each replicon variant. Error bars represent the standard errors (SEs) of three independent biological replicates (n = 6 wheat calli per replicate) transformed by particle bombardment. Gold particles with no DNA were used to transform the wild-type (WT) control.
