Abstract
Abstract
Nucleic acid therapeutics offer a new paradigm to rapidly respond to global health problems. The versatility of nucleic acids, especially in RNA therapies, provides the ability to tune levels of specific protein expression, achieving downregulation through short interfering RNA (siRNA) or upregulation by messenger RNA (mRNA) administration. Recent advances in the development of delivery vehicles, including nonviral nanoparticles are crucial to overcome the innate barriers to nucleic acid delivery. Toward this end, current clinical approaches have utilized mRNA and lipid nanoparticles (LNPs) to address the COVID-19 pandemic through novel vaccine strategies, producing efficacious vaccines within one year of sequencing the SARS-CoV-2 genome. Here, we review fundamental concepts required to achieve successful nucleic acid delivery, including the design of LNP systems optimized for mRNA vaccine applications.
Graphic abstract
Keywords: Nanostructures, RNA therapeutics, Lipid nanoparticles, COVID-19, Vaccines
Introduction
Nucleic acid therapeutics represent a promising new paradigm of treatment, which can prevent or treat the root causes of human disease by harnessing the potential of the genetic code.1 Indeed, there are near infinite therapeutic applications for nucleic acid drugs due to the diversity of protein targets. For example, short interfering RNAs (siRNAs) and antisense oligonucleotides (ASOs) can be tailored to knockdown (i.e., decrease) levels of a harmful mutant protein in cancers considered undruggable by small molecules. Rare genetic diseases can be treated with the replacement of mutated genes using delivered DNA or messenger RNA (mRNA). As evidenced by the efforts to overcome the global COVID-19 pandemic, mRNA can be implemented to produce therapeutic proteins or train the immune system as novel vaccines.2 Until recently, however, several challenges, including the inefficient delivery of naked nucleic acids prevented the widespread development and clinical approval of nucleic acid-based drugs.3 In this way, the development of safe and effective carrier systems, including nonviral nanoparticles, has become paramount to the translational success of nucleic acid drugs, including innovative COVID-19 mRNA vaccines.4
In this article, we will provide an overview of the fundamental background research that has helped to guide the current development of nonviral lipid nanoparticles (LNPs) for use as mRNA vaccines. Specifically, we will outline the generalizable therapeutic paradigms that may be accessed through RNA-based drugs, including downregulation and induced protein expression. We will further focus on the key biological barriers that prevent the successful delivery of naked nucleic acids, including serum instability, immunogenicity, and poor cellular uptake. Finally, we will explore the design, synthesis, and evaluation of nonviral nanocarriers such as LNPs, which have been clinically deployed to hundreds of millions of patients in current COVID-19 mRNA vaccines. This article will provide fundamental insights into the development of nucleic acid drugs as well as more broadly delineate the state of the art in the implementation of nonviral nanoparticle delivery vehicles for translational applications.
Therapeutic nucleic acids
Countless diseases are caused by aberrations in protein expression. In some instances, a mutation results in translation of a toxic or harmful protein that must be knocked down. Alternatively, some null mutations prevent or reduce the expression of a vital protein, necessitating the upregulation or replacement of this gene. In all cases, the underlying cellular dysfunction can be addressed using various forms of therapeutic nucleic acids.5 Of particular therapeutic interest is RNA, the crucial intermediate between genome and protein. Unlike DNA, which must reach the nucleus to be transcribed, RNA need only enter the cytosol to be translated to protein.6 RNA is typically composed of the canonical Watson–Crick nucleotide bases: adenine (A), cytosine (C), guanine (G), and uracil (U). Thus, unlike proteins or small molecules that can exhibit extreme variety in their surface properties and structure, the shared code of nucleic acid bases provides overarching similarities in their physical properties. RNA is transiently expressed due to the presence of the 2ʹ-hydroxyl group of the ribose sugar which facilitates hydrolysis and enzymatic degradation, providing temporal control of RNA therapeutics. Currently, the most clinically advanced therapies using nonviral delivery vectors encapsulate therapeutic siRNA or mRNA.
The discovery of siRNA in 1998 in Caenorhabditis elegans identified a new molecular tool capable of specific protein knockdown.7 Exogenous therapeutic siRNA is typically administered as a double-stranded, 21–24 nucleotide (nt) construct, with overhangs formed through cleavage by an enzyme called Dicer.8 The antisense strand of the siRNA is loaded into the RNA-induced silencing complex (RISC) with recruited Argonaute proteins (Ago), which enables specific targeting of the sense sequence in an mRNA transcript.9 The RISC subsequently cleaves and degrades the target mRNA through the RNase activity of Ago, thereby preventing protein translation.
Alternatively, mRNA can be used to induce expression of a therapeutic protein as first demonstrated in vivo by Wolff et al. through intramuscular (IM) injections of mRNA in mice.10 mRNA is a single-stranded construct, often thousands of nt in length encoding for a protein of interest. Eukaryotic mRNA also contains several natural modifications to promote ribosomal recognition and translation. After spliceosomal processing and removal of noncoding introns, the remaining exons which encode for a given gene are left along with a 5ʹ and 3ʹ untranslated region (UTR) on either side of this coding sequence.11 These UTR help to regulate the overall level of translation as well as the stability of the transcript. Native mRNA transcribed within cells exhibits a 7-methylguanosine (m7G) cap comprised of the modified nucleotide which is enzymatically joined in a 5ʹ–5ʹ bond and aids in ribosomal binding. Finally, the mRNA transcript is appended with a poly-adenylate (polyA) tail, which reduces exonuclease degradation and recruits polyA-binding proteins (PABP) in translation. A desired mRNA transcript can be synthesized using commercially available enzymes to include not only the open reading frame (orf) of a gene of interest, but also to include these crucial modifications.12 This process of in vitro transcription (IVT) is also readily scalable to increase the output of the desired mRNA.
Delivery barriers and nucleic acid modifications
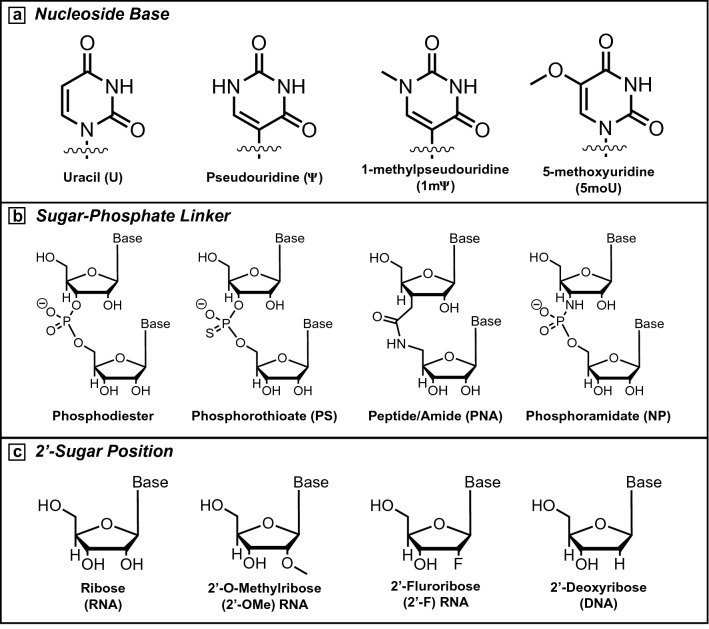
Although the therapeutic applications of nucleic acids have long been recognized, several delivery barriers must be overcome to achieve this potential. At the cellular level, naked RNA is vulnerable to degradation as the innate immune system is sensitive to foreign RNA presented by viral and bacterial pathogens. These barriers include pattern-recognition receptors (PRR) activated by naked RNA as a pathogen-associated molecular pattern (PAMP).13 Single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) both activate Toll-like Receptors (TLR) as part of the innate immune system leading to inflammation and heightened immunogenicity.14,15 To evade these responses, researchers often incorporate non-canonical, chemically modified nucleoside bases outside of the Watson–Crick bases (A, G, C, U) (Figure 1a). Karikó et al. reported that the replacement of uracil with pseudouridine (Ψ) in an mRNA sequence resulted in reduced immune activation while also improving translation of the desired protein.16 Additional research evaluating further modifications, including 1-methylpseudouridine (1mΨ) and 5-methoxyuridine (5moU) indicated cell-type differences in protein expression but also found improved sequence translation and overall stability.17 Removal of dsRNA impurities can be achieved either through IVT process optimization or through high performance liquid chromatography (HPLC).18,19 These purifications can reduce undesired immune activation and improve overall delivery efficacy, although the reactogenicity caused by nucleic acids can be beneficial in activating the immune response for certain vaccine technologies.
Figure 1.
Common modifications to nucleic acids. (a) Uracil is often modified to pseudouridine (ψ), 1-methylpseudouridine (1mψ), or 5-methoxyuridine (5moU) to evade immune activation and improve translation in mRNA sequences. (b) Alternative linkages are used to improve stability of the sugar-phosphate backbone, including phopshorothioate (PS), peptide (PNA), or phosphoramidate linkages (NP). (c) The 2ʹ-hydroxyl position native to the ribose sugar is often modified to reduce the incidence rate of hydrolysis and promote sequence stability. Common modifications include 2ʹ-O-methyl (2ʹ-OMe) and 2ʹ-Fluoro (2ʹ-F) in addition to naturally occurring 2ʹ-deoxyribose sugars (DNA).
Beyond the cellular immune response, the presence of RNases and endonucleases within the serum can degrade RNA when delivered systemically or intravenously (IV).20 To avoid nuclease degradation, sequences incorporating alternative linker chemistries or changes to the 2ʹ-position of the furanose sugar have been developed (Figure 1b–c).21 These modifications are more common in siRNA but are rarely found in mRNA sequences, as the backbone modifications often sterically interfere with effective translation. IV injections of RNA or nanoparticle delivery vehicles must also bypass the reticuloendothelial system (RES) and fenestrae within the liver, which selectively uptakes particles in the 80–200-nm size range.22 Nucleic acids are large molecules and are strongly negatively charged due to the sugar-phosphate backbone. As a result of their size and charge, naked RNA cannot freely cross the cellular membrane, compelling the need for delivery vehicles, which not only protect RNA from degradation, but also facilitate intracellular uptake.23
Nonviral delivery vectors and lipid nanoparticles
Recent efforts have expanded the use of nonviral nanoparticle vectors to protect RNA cargo and deliver to specific cell populations. Although viruses act as potent vectors for cellular transfection, their use has been reviewed extensively elsewhere.24 Early nanoparticle strategies utilized polymers and lipids with cationic quaternary amines (such as polyethyleneimine (PEI) or dioleoyl trimethylammonium propane (DOTAP)) to electrostatically complex the anionic nucleic acid backbone into polyplexes or lipoplexes.25 However, the resulting particles often exhibit a strong surface charge, which can lead to unwanted toxicity and reduced efficacy in vivo. To avoid these properties, researchers have shifted their focus to multicomponent LNP utilizing synthetic ionizable lipids with tertiary amine groups to maintain a more neutral charge. Several nanoparticle formulation methods have been developed including lipid film hydration and bulk mixing. However, microfluidic mixing is most commonly used as good manufacturing practice (GMP) as both a scalable and replicable method to produce consistent laboratory- or industrial-scale batches of LNP.26
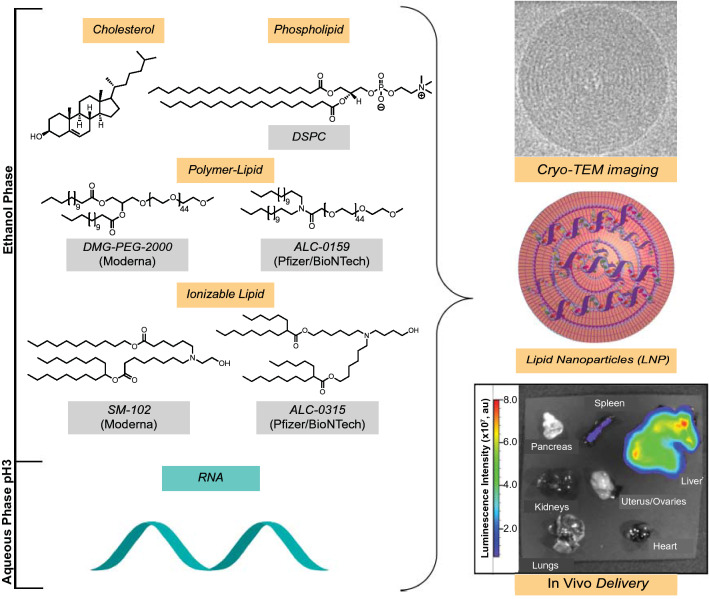
LNP are typically formulated through a combination of four chemical components: phospholipid, cholesterol, lipid-anchored polymers, and an ionizable lipid (Figure 2).27,28 Phospholipids [e.g., distearoyl phosphocholine (DSPC)] and cholesterol help to form and stabilize a lipid bilayer encapsulating the nucleic acid cargo. Lipid-anchored polymers typically contain hydrocarbon lipid tails and long poly(ethylene glycol) (PEG) polymeric chains that prevent the nonspecific uptake of nanoparticles to increase circulation time. The ionizable lipid component is considered essential, not only for complexation and encapsulation of the RNA cargo, but also for aiding in endosomal escape and intracellular delivery. LNP can be characterized using dynamic light scattering (DLS) or cryo-TEM to determine particle size and overall surface charge. Using fluorescently tagged-RNA or mRNA encoding for fluorescent (such as GFP) or bioluminescent proteins (such as firefly luciferase), the biodistribution of LNP delivery and the expression kinetics in various organs can be assessed (Figure 2).29,30 In 2018, Alnylam Pharmaceuticals received the first FDA approval for an siRNA LNP drug for Onpattro, which uses an ionizable lipid component (DLin-MC3-DMA or MC3) to encapsulate siRNA encoding against transthyretin (TTR) protein to treat TTR-mediated amyloidosis in the liver.31
Figure 2.
Components used in lipid nanoparticle (LNP) formulation. Cholesterol, phospholipids, PEG-lipid, and ionizable lipid are combined in ethanol or organic solvent while RNA is dissolved in an aqueous buffer. Microfluidic mixing methods can yield LNP encapsulating therapeutic RNA cargo. LNP can be characterized by cryo-transmission electron microscopy (cryo-TEM), showing a spherical morphology. In vivo protein expression after IV administration of LNP encapsulating mRNA encoding for firefly luciferase (a light-emitting protein) is predominantly observed in the liver.29 DSPC, distearoyl phosphocholine; DMG, dimyristoyl glycerols; PEG, poly(ethylene glycol).
While IV administration typically results in delivery to the liver, we have previously identified novel ionizable lipids, which when incorporated in LNP, result in delivery to other organs of interest such as the spleen.32 LNP formulations with certain ionizable lipids have been engineered to achieve preferential delivery to B lymphocytes in the spleen or ex vivo T-cells.33,34 Synergistic co-formulations of ionizable lipids and specific helper lipids can also be tailored to achieve delivery in the liver, spleen, or lungs.35,36 Similarly, Oberli et al. screened several iterative libraries of LNP formulations to identify an optimized formulation for cancer immunotherapy, with the lead formulation capable of transfecting dendritic cells (DCs) in the inguinal lymph nodes to activate the immune system.37 Hassett et al. also screened a library of ionizable lipids to identify a lead candidate for IM administration to transfect cells within the lymph nodes as potential mRNA vaccines. Their selected lipid, SM-102 (Figure 2, discussed next), produced immune titers comparable to clinically approved MC3 and considerably improved biodegradability and tolerability, providing a platform for repeated LNP dosing.38
Although the underlying mechanisms of intracellular delivery remain unresolved, several theories have been suggested and explored in the literature. LNP are typically brought into the cell through endocytosis, often through receptor-mediated interactions from the protein corona.39 Within the endosome, fusogenic lipids, including the phospholipid dioleoyl phosphoethanolamine (DOPE) and certain ionizable lipids, are hypothesized to form an inverted hexagonal bilayer structure (HII), which disrupts the endosomal membrane to release the internal RNA cargo.40 Current research suggests that endosomal escape remains the rate-limiting step for RNA delivery, as only ~ 2–10% of LNP release their cargo to the cytosol.41 Despite the relatively low success of RNA cargo in reaching the ultimate intracellular goal, RNA therapies act in a catalytic fashion, such that one copy of siRNA or mRNA can be repeatedly used in the inhibition or initiation of translation, respectively. Therefore, delivery of few therapeutic RNA can have a substantial impact on the protein levels within the cell or tissue. Importantly, delivery vehicles such as LNP are generalizable and can be applied to RNA for various therapeutic applications. While formulation optimization may be necessary when comparing siRNA or mRNA, or when targeting specific organs as mentioned above, the similarities in physical properties of nucleic acids allow for efficacious delivery regardless of the specific sequence to be encapsulated.42 Thus, LNP can be considered platform technologies, aiding development and deployment to treat a myriad of disorders, including the use of nucleic acids as potential vaccines against infectious disease or cancers.
Design of mRNA vaccines to combat COVID-19
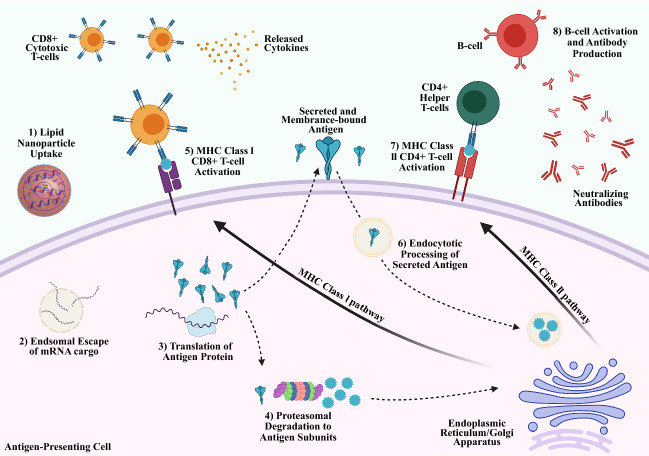
Emerging research interest has focused on the use of mRNA to inoculate patients against infectious diseases, first evaluated by Martinon et al. in 1993 using loaded liposomes to immunize mice against influenza.43 Rather than traditional vaccines that include a specific protein component of a virus, or even an attenuated version of the virus itself, nucleic acid vaccines instead deliver the genetic code to express a viral protein or antigen. The full scope of the immunological response in vaccination to infectious diseases is beyond the scope of this review and has been reviewed elsewhere.44,45 Current hypotheses hold that delivery of mRNA cargo encoding for an antigen protein to DCs can spur the maturation into antigen-presenting cells (APCs) to train the immune system to produce antibodies (Abs) recognizing this antigen target (Figure 3).46–48 Uptake and expression of the antigen-encoding mRNA results in endogenous production of the “foreign” antigen within DCs. DCs may then mature into APCs with the antigen protein present on the cellular surface to activate the cellular and humoral immune system against the pathogen. Alternatively, proteasomal degradation of the antigen protein can result in certain motifs or subunits entering the major histocompatibility complex (MHC) class I pathway, whereby the antigen becomes present to CD8+T-cells.49 These cytotoxic T-cells are activated by the MHC class I receptor and co-immunostimulatory ligands. Mature CD8+T-cells can then secrete immune-activating cytokines and may also induce apoptosis in infected or mutated cells presenting the detected antigen. Humoral immunity can be developed through a similar process with endocytosis of secreted or intact antigen proteins. These proteins are degraded and processed through the MHC class II pathway, where antigen molecules are presented to CD4+T-cells.50 These helper T-cells become activated and subsequently train B-cells which can produce large quantities of antibodies against the antigen with the potential to establish long-term memory in B-cells as well.51 This strategy of mRNA vaccination and immune activation has been applied in research investigating not only infectious diseases, but also developing cancer immunotherapies. Prior to 2020, despite promising preclinical results no mRNA vaccine candidate had successfully advanced through clinical trials.
Figure 3.
Proposed immune activation pathways for mRNA vaccines. Lipid nanoparticles with mRNA encoding an antigen protein are injected intramuscularly and uptaken by dendritic cells. mRNA cargo undergoes endosomal escape to be translated into the full or partial antigen protein by ribosomes. The antigen protein may be expressed on the surface of antigen-presenting cells (APCs), may be degraded into peptide subunits major histocompability complex ([MHC] class I pathway), or may be secreted and may subsequently enter other APCs (MHC Class II pathway). When presented by MHC class I, the peptide complex will activate CD8+Cytotoxic T-cells against the antigen with subsequent cytokine release and cellular-induced immunity. When presented by MHC class II, the antigen peptide will activate CD4+ or Helper T-cells, leading to humoral immunity and neutralizing antibody production toward the antigen.46
As the COVID-19 pandemic caused by the SARS-CoV-2 virus spread globally, mRNA vaccines emerged as the technology which could most rapidly respond to the unmet need to immunize the world population. The RNA genome of this coronavirus was synthesized in early 2020 with researchers identifying the viral spike protein as crucial to cellular entry through interactions with the ACE2 receptor of the host cells.52 With knowledge of the viral genome and utilizing an LNP delivery strategy, industrial groups, including Moderna Therapeutics (Cambridge, MA, USA) and BioNTech (partnered with Pfizer for global production and distribution; Mainz, Germany) have developed vaccine platforms that produce Abs against a perfusion stabilized form of the spike protein on the SARS-CoV-2 viral surface. These vaccines received Emergency Use Authorization from the FDA in 2020 and are currently the only mRNA therapeutics in use in patients. In August 2021, the Pfizer/BioNTech COVID-19 vaccine (under the brand name Comirnaty) received full FDA approval.53
Moderna, in partnership with the National Institute of Allergy and Infectious Diseases (NIAID), was able to rapidly develop their vaccine candidate, mRNA-1273, with modified mRNA encapsulated in LNP formulated with SM-102, cholesterol, DSPC, and DMG-PEG2000 (PEG-lipid) (Figure 2). Preclinical studies showed strong levels of Ab production and protective immunity in non-human primates (NHP) with high levels of CD4+T-cell activation, although with low observed CD8+T-cell activation.54 After passing phase I/II safety trials, the vaccine was swiftly brought to large-scale phase III clinical trials to evaluate overall efficacy, with patients receiving a 100-µg dose of mRNA-1273 administered twice over 28 days.55 Across nearly 30,000 total participants matched 1:1 with placebo doses (n = 14,134 treatment versus n = 14,073 placebo), the vaccine regimen showed a 94.1% efficacy (CI 89.3–98.6%). Further patient analysis found that levels of CD4+ and CD8+T-cell immunity replicated preclinical observations. In trial groups, there were 11 identified cases in patients receiving the vaccine and 185 cases in those receiving placebo vaccines. Promisingly, a small cohort of vaccinated patients retain neutralizing levels of Ab production at least six months post-vaccination (although as of the time of this writing, longer timepoints have not yet been reached).56 Initial research indicates a decrease in efficacy against certain viral mutations; however, the overall neutralizing effect of Abs remains above the desired threshold.57
BioNTech developed their vaccine candidate BNT162b2 as a modified mRNA encapsulated within an LNP composed of the ionizable lipid ALC-0315, cholesterol, DSPC, and PEG-lipid ALC-0159 (Figure 2). Early clinical trials showed that the induced Ab production reached levels greater than that observed in convalescent plasma of COVID-19 patients upon a 30-µg dose delivered twice over 21 days.58 In smaller observation cohorts, the majority of patients showed an orders of magnitude level of increase in the presence of active CD4+ and CD8+T-cells after the second-dose boost in both the BNT162b2 and a similar vaccine BNT162b1 which encodes for a secreted version of the spike protein.59,60 Across a larger phase III study of more than 40,000 patients matched 1:1 with placebo controls (n = 18,198 treatment versus n = 18,325 placebo), the BNT162b2 vaccine was shown to be 95% effective (CI 90.0–97.9%) in preventing COVID-19 infection.61 There were eight observed cases in vaccinated patients compared to 162 cases in the placebo groups. Interim analyses also indicate that neutralizing Abs are effective against several circulating variants of SARS-CoV-2, although patients will continue to be monitored long term.62 Booster shots of mRNA vaccines are being investigated and recommended for certain patients including the elderly and immuno compromised. Recent analyses have detected a decrease in antibody titers over time, however, the vaccines remain strongly efficacious in protecting against severe disease outcomes (97% or higher reduction in hospitalizations and deaths).63–65
Outlook and future potential
The COVID-19 pandemic may ultimately serve as the catalyst to usher in a new wave of nucleic acid therapeutics. The rapid deployment of mRNA vaccines was made possible by decades of prior research focused on the development of LNP as a delivery vehicle for therapeutic RNA. Now, with hundreds of millions of patients receiving these treatments, mRNA vaccines are proving real-world efficacy in preventing hospitalization and death from serious infectious disease. Crucially, the LNP platform can be adapted to provide booster doses or adaptive vaccines in the face of viral mutations which may escape existing immunity. Novel mRNA transcripts can be generated to respond to the evolution of the SARS-CoV-2 virus or the emergence of future pathogens. By utilizing the same LNP technology, full-scale clinical trials may not be necessary or can be rapidly expedited to combat variants as they arise. With these promising results, researchers can further expand the uses of LNP to address other threats to global health, including cancer through vaccine-mediated immunotherapy. With potent LNP delivery vectors in hand, novel RNA sequences can be used to induce or knockdown protein expression within a variety of target cell populations. Additional improvements in ionizable lipid design, RNA technology (such as self-amplifying mRNA which requires less material), and industrial manufacturing may soon lead to widespread benefits of RNA delivery.
Acknowledgments
Figures created with Biorender.com. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Biographies
Jason L. Andresen
is a doctoral candidate in the Department of Chemistry at the Massachusetts Institute of Technology, currently conducting research in the laboratory of Professor Robert Langer. He received his BS degree in biochemistry and molecular biology from the University of Richmond in 2015. His research focuses on novel uses of lipid nanoparticles for RNA delivery to address currently untreatable conditions. Previously, he conducted research on the synthesis of hydrogel biomaterials for drug delivery and tissue-engineering applications. He can be reached by email at jandrese@mit.edu.
Owen S. Fenton
is an assistant professor in the Department of Chemical and Biomolecular Engineering at the National University of Singapore. Previously, Owen studied in the laboratories of Professors Robert Langer and Daniel Anderson at the Massachusetts Institute of Technology, completing his PhD studies in 2016. His research group focuses on the molecular engineering of synthetic drug delivery systems with a particular focus on the development of advanced RNA therapeutics for the study, prevention, and treatment of disease. He can be reached by email at cheosf@nus.edu.sg.
Contributor Information
Jason L. Andresen, Email: jandrese@mit.edu
Owen S. Fenton, Email: cheosf@nus.edu.sg
References
- 1.Singh MS, Peer D. RNA nanomedicines: The next generation drugs? Curr. Opin. Biotechnol. 2016;39:28. doi: 10.1016/j.copbio.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Schlake T, Thess A, Fotin-Mleczek M, Kallen KJ. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kauffman KJ, Webber MJ, Anderson DG. Materials for nonviral intracellular delivery of messenger RNA therapeutics. J. Control. Release. 2016;240:227. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 4.Moss KH, Popova P, Hadrup SR, Astakhova K, Taskova M. Lipid nanoparticles for delivery of therapeutic RNA oligonucleotides. Mol. Pharm. 2019;16:2265. doi: 10.1021/acs.molpharmaceut.8b01290. [DOI] [PubMed] [Google Scholar]
- 5.Fenton OS, Olafson KN, Pillai PS, Mitchell MJ, Langer R. Advances in biomaterials for drug delivery. Adv. Mater. 2018 doi: 10.1002/adma.201705328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Y, Siegwart DJ, Anderson DG. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019;144:133. doi: 10.1016/j.addr.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 11.Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 12.Kwon H, Kim M, Seo Y, Moon YS, Lee HJ, Lee K, Lee H. Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials. 2018;156:172. doi: 10.1016/j.biomaterials.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 13.C. A. Janeway, Approaching the asymptote? Evolution and revolution in immunology, in Cold Spring Harbor Symposium on Quantitative Biology (Cold Spring Harbor, NY, Laboratory Press, 1989), pp. 1–13. [DOI] [PubMed]
- 14.Lokugamage MP, Gan Z, Zurla C, Levin J, Islam FZ, Kalathoor S, Sato M, Sago CD, Santangelo PJ, Dahlman JE. Mild innate immune activation overrides efficient nanoparticle-mediated RNA delivery. Adv. Mater. 2020;32:1904905. doi: 10.1002/adma.201904905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for toll-like receptor 3. J. Biol. Chem. 2004;279:12542. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 16.Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Luo X, Dong Y. Effects of chemically modified messenger RNA on protein expression. Bioconjug. Chem. 2016;27:849. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- 18.Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson J, Sorensen EW, Mintri S, Rabideau AE, Zheng W, Besin G, Khatwani N, Su SV, Miracco EJ, Issa WJ, Hoge S, Stanton MG, Joyal JL. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brad Wan W, Seth PP. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 2016;59:9645. doi: 10.1021/acs.jmedchem.6b00551. [DOI] [PubMed] [Google Scholar]
- 22.Mitragotri S, Burke PA, Langer R. Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Rev. Drug Discov. 2014;13:655. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 24.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: Progress and challenges. Nat. Rev. Genet. 2011;12:341. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 25.Hajj KA, Whitehead KA. Tools for translation: Nonviral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 26.E. Thomée, Microfluidic nanoparticle synthesis: A short review, Elveflow. (2017). https://www.elveflow.com/microfluidic-reviews/general-microfluidics/microfluidic-nanoparticle-synthesis-short-review. Accessed 20 Mar 2021
- 27.Levins CG, Langer R, Querbes W, Whitehad KA, Alvarez R, Racie T, Qin LL, Love KT, Yip KN, Dorkin JR, Cantley W, Koteliansky V, Mahon KP, Fitzgerald K, Qin J, de Fougerolles A, Sah DWY, Anderson DG, Frank-Kamenetsky M, Akinc A. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1864. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkarni JA, Witzigmann D, Leung J, van der Meel R, Zaifman J, Darjuan MM, Grisch-Chan HM, Thöny B, Tam YYC, Cullis PR. Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale. 2019;11:9023. doi: 10.1039/C9NR02004G. [DOI] [PubMed] [Google Scholar]
- 29.Fenton OS, Kauffman KJ, McClellan RL, Appel EA, Dorkin JR, Tibbitt MW, Heartlein MW, DeRosa F, Langer R, Anderson DG. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater. 2016;28:2939. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardi N, Tuyishime S, Muramatsu H, Karikó K, Mui BL, Tam YK, Madden TD, Hope MJ, Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, Ansell S, Du X, Hope MJ, Madden TD, Mui BL, Semple SC, Tam YK, Ciufolini M, Witzigmann D, Kulkarni JA, van der Meel R, Cullis PR. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 32.Fenton OS, Kauffman KJ, McClellan RL, Kaczmarek JC, Zeng MD, Andresen JL, Rhym LH, Heartlein MW, DeRosa F, Anderson DG. Customizable lipid nanoparticle materials for the delivery of siRNAs and mRNAs. Angew. Chem. Int. Ed. 2018;57:13582. doi: 10.1002/anie.201809056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Billingsley MM, Singh N, Ravikumar P, Zhang R, June CH, Mitchell MJ. Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering. Nano Lett. 2020;20:1578. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenton OS, Kauffman KJ, Kaczmarek JC, McClellan RL, Jhunjhunwala S, Tibbitt MW, Zeng MD, Appel EA, Dorkin JR, Mir FF, Yang JH, Oberli MA, Heartlein MW, DeRosa F, Langer R, Anderson DG. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater. 2017;29:1606944. doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020 doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao L, Lin J, Huang Y, Li L, Delcassian D, Ge Y, Shi Y, Anderson DG. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020 doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberli MA, Reichmuth AM, Dorkin JR, Mitchell MJ, Fenton OS, Jaklenec A, Anderson DG, Langer R, Blankschtein D. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassett KJ, Benenato KE, Jacquinet E, Lee A, Woods A, Yuzhakov O, Himansu S, Deterling J, Geilich BM, Ketova T, Mihai C, Lynn A, McFadyen I, Moore MJ, Senn JJ, Stanton MG, Almarsson Ö, Ciaramella G, Brito LA. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AKR, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3955. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel S, Ashwanikumar N, Robinson E, Xia Y, Mihai C, Griffith JP, Hou S, Esposito AA, Ketova T, Welsher K, Joyal JL, Almarsson Ö, Sahay G. Naturally occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020 doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K, Manoharan M, Kirchhausen T, Lieberman J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, Fenton OS, Anderson DG. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 43.Martinon F, Krishnan S, Lenzen G, Magné R, Gomard E, Guillet JGG, Lévy JPP, Meulien P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23:1719. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 44.Plotkin SA. Correlates of vaccine-induced immunity. Clin. Infect. Dis. 2008;47:401. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 45.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12:509. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wadhwa A, Aljabbari A, Lokras A, Foged C, Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines-A new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reichmuth AM, Oberli MA, Jaklenec A, Langer R, Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Midoux P, Pichon C. Lipid-based mRNA vaccine delivery systems. Informa Healthcare. 2014 doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 50.Lindgren G, Ols S, Liang F, Thompson EA, Lin A, Hellgren F, Bahl K, John S, Yuzhakov O, Hassett KJ, Brito LA, Salter H, Ciaramella G, Loré K. Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pardi N, Hogan MJ, Naradikian MS, Parkhouse K, Cain DW, Jones L, Moody MA, Verkerke HP, Myles A, Willis E, LaBranche CC, Montefiori DC, Lobby JL, Saunders KO, Liao HX, Korber BT, Sutherland LL, Scearce RM, Hraber PT, Tombácz I, Muramatsu H, Ni H, Balikov DA, Li C, Mui BL, Tam YK, Krammer F, Karikó K, Polacino P, Eisenlohr LC, Madden TD, Hope MJ, Lewis MG, Lee KK, Hu SL, Hensley SE, Cancro MP, Haynes BF, Weissman D. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.FDA, FDA Approves First COVID-19 Vaccine. (FDA Press Release, 2021). https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine. Accessed 25 Aug 2021.
- 54.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, Nagata BM, Andersen H, Martinez DR, Noe AT, Douek N, Donaldson MM, Nji NN, Alvarado GS, Edwards DK, Flebbe DR, Lamb E, Doria-Rose NA, Lin BC, Louder MK, O’Dell S, Schmidt SD, Phung E, Chang LA, Yap C, Todd J-PM, Pessaint L, Van Ry A, Browne S, Greenhouse J, Putman-Taylor T, Strasbaugh A, Campbell T-A, Cook A, Dodson A, Steingrebe K, Shi W, Zhang Y, Abiona OM, Wang L, Pegu A, Yang ES, Leung K, Zhou T, Teng I-T, Widge A, Gordon I, Novik L, Gillespie RA, Loomis RJ, Moliva JI, Stewart-Jones G, Himansu S, Kong W-P, Nason MC, Morabito KM, Ruckwardt TJ, Ledgerwood JE, Gaudinski MR, Kwong PD, Mascola JR, Carfi A, Lewis MG, Baric RS, McDermott A, Moore IN, Sullivan NJ, Roederer M, Seder RA, Graham BS. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doria-Rose N, Suthar MS, Makowski M, O’Connell S, McDermott AB, Flach B, Ledgerwood JE, Mascola JR, Graham BS, Lin BC, O’Dell S, Schmidt SD, Widge AT, Edara V-V, Anderson EJ, Lai L, Floyd K, Rouphael NG, Zarnitsyna V, Roberts PC, Makhene M, Buchanan W, Luke CJ, Beigel JH, Jackson LA, Neuzil KM, Bennett H, Leav B, Albert J, Kunwar P. Antibody persistence through 6 months after the second dose of mRNA-1273 vaccine for COVID-19. N. Engl. J. Med. 2021 doi: 10.1056/nejmc2103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu K, Werner AP, Koch M, Choi A, Narayanan E, Stewart-Jones GBE, Colpitts T, Bennett H, Boyoglu-Barnum S, Shi W, Moliva JI, Sullivan NJ, Graham BS, Carfi A, Corbett KS, Seder RA, Edwards DK. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Lyke KE, Raabe V, Dormitzer PR, Jansen KU, Sahin U, Gruber WC. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sahin U, Muik A, Vogler I, Derhovanessian E, Kranz LM, Vormehr M, Quandt J, Bidmon N, Ulges A, Baum A, Pascal K, Maurus D, Brachtendorf S, Lörks V, Sikorski J, Koch P, Hilker R, Becker D, Eller AK, Grützner J, Tonigold M, Boesler C, Rosenbaum C, Heesen L, Kühnle MC, Poran A, Dong JZ, Luxemburger U, Kemmer-Brück A, Langer D, Bexon M, Bolte S, Palanche T, Schultz A, Baumann S, Mahiny AJ, Boros G, Reinholz J, Szabó GT, Karikó K, Shi PY, Fontes-Garfias C, Perez JL, Cutler M, Cooper D, Kyratsous CA, Dormitzer PR, Jansen KU, Türeci Ö. BNT162b2 induces SARS-CoV-2-neutralizing antibodies and T cells in humans. MedRxiv. 2020 doi: 10.1101/2020.12.09.20245175. [DOI] [Google Scholar]
- 60.Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, Baum A, Pascal K, Quandt J, Maurus D, Brachtendorf S, Lörks V, Sikorski J, Hilker R, Becker D, Eller AK, Grützner J, Boesler C, Rosenbaum C, Kühnle MC, Luxemburger U, Kemmer-Brück A, Langer D, Bexon M, Bolte S, Karikó K, Palanche T, Fischer B, Schultz A, Shi PY, Fontes-Garfias C, Perez JL, Swanson KA, Loschko J, Scully IL, Cutler M, Kalina W, Kyratsous CA, Cooper D, Dormitzer PR, Jansen KU, Türeci Ö. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 61.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Sahin U, Jansen KU, Gruber WC. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020;383:2603. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, Liu J, Xia H, Zhang X, Fontes-Garfias CR, Swanson KA, Cai H, Sarkar R, Chen W, Cutler M, Cooper D, Weaver SC, Muik A, Sahin U, Jansen KU, Xie X, Dormitzer PR, Shi P-Y. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384:1466. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Callaway E. COVID vaccine boosters: the most important questions. Nature. 2021;596:178–180. doi: 10.1038/d41586-021-02158-6. [DOI] [PubMed] [Google Scholar]
- 64.Kamar N, Abravanel F, Marion O, Couat C, Izopet J, Del Bello A. Three doses of an mRNA Covid-19 vaccine in solid-organ transplant recipients. N. Engl. J. Med. 2021;385:661–662. doi: 10.1056/nejmc2108861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.B. Mizrahi, R. Lotan, N. Kalkstein, A. Peretz, G. Perez, A. Ben-Tov, G. Chodick, S. Gazit, T. Patalon, Correlation of SARS-CoV-2 breakthrough infections to time-fromvaccine; Preliminary Study. MedRxiv (2021). 10.1101/2021.07.29.21261317 [DOI] [PMC free article] [PubMed]