Abstract
BACKGROUND
Germline mutations in telomerase and other telomere maintenance genes manifest in the premature aging short telomere syndromes. Myelodysplastic syndromes and acute myeloid leukemia (MDS/AML) account for 75% of associated malignancies, but how these cancers overcome the inherited telomere defect is unknown.
METHODS
We used ultra-deep targeted sequencing to detect somatic reversion mutations in 17 candidate telomere lengthening genes among controls and patients with short telomere syndromes with and without MDS/AML, and we tested the functional significance of these mutations.
RESULTS
While no controls carried somatic mutations in telomere maintenance genes, 29% (16 of 56) of adults with germline telomere maintenance defects carried at least 1 (P < 0.001), and 13% (7 of 56) had 2 or more. In addition to TERT promoter mutations, which were present in 19%, another 13% of patients carried a mutation in POT1 or TERF2IP. POT1 mutations impaired telomere binding in vitro and some mutations were identical to ones seen in familial melanoma associated with longer telomere length. Exclusively in patients with germline defects in telomerase RNA (TR), we identified somatic mutations in nuclear RNA exosome genes RBM7, SKIV2L2, and DIS3, where loss-of-function upregulates mature TR levels. Somatic reversion events in 6 telomere-related genes were more prevalent in patients who were MDS/AML-free (P = 0.02, RR 4.4, 95% CI 1.2–16.7), and no patient with MDS/AML had more than 1 reversion mutation.
CONCLUSION
Our data indicate that diverse adaptive somatic mutations arise in the short telomere syndromes. Their presence may alleviate the telomere crisis that promotes transformation to MDS/AML.
FUNDING
This work was supported by the NIH, the Commonwealth Foundation, the S&R Foundation Kuno Award, the Williams Foundation, the Vera and Joseph Dresner Foundation, the MacMillan Pathway to Independence Award, the American Society of Hematology Scholar Award, the Johns Hopkins Research Program for Medical Students, and the Turock Scholars Fund.
Keywords: Genetics, Oncology
Keywords: Clonal selection, Leukemias, Telomeres
Introduction
Germline mutations in telomerase and other telomere-related genes are the most common known genetic cause of adult-onset myelodysplastic syndromes and possibly also acute myeloid leukemia (MDS/AML) (1–4). In line with this observation, MDS/AML are the most common cancers in patients with inherited mutations in telomerase and other telomere maintenance genes (5, 6). They account for 75% of short telomere syndrome malignancies, although the lifetime risk is 10% (5, 6). The vast majority of short telomere syndrome MDS/AML cases are age-related; they manifest after the age of 50, often concurrently with idiopathic pulmonary fibrosis or other telomere-mediated pulmonary disease (5–8). In primary cells, critically short telomeres provoke a DNA damage response, which triggers a p53-dependent checkpoint that signals apoptosis or cellular senescence (9, 10). This checkpoint mediates the degenerative phenotypes of the short telomere syndromes, which manifest clinically as bone marrow failure and pulmonary fibrosis (11, 12). How these myeloid cancers overcome the background short telomere checkpoint to sustain their replicative potential remains unknown. Moreover, the factors that predict which patients go on to develop MDS/AML are not understood. The latter question is particularly timely, since pulmonary fibrosis patients with short telomere syndromes are now identifiable prior to lung transplantation, but have a significantly increased risk of acquiring posttransplant MDS and AML (6).
Here, we used an ultra-deep sequencing approach to test the hypothesis that acquired clonal somatic mutations that offset the inherited short telomere defect arise in telomere-mediated MDS/AML. We found diverse somatic reversion mechanisms that have great specificity relative to the germline defect. In contrast to our expectations, however, these somatic mutations were rare in patients with MDS/AML and their allele burden was significantly higher in patients who were MDS/AML-free. These data have implications for fundamental understanding of how somatic adaptation in the setting of defective hematopoiesis may provide protection against MDS/AML evolution in at least some inherited bone marrow failure syndromes.
Results
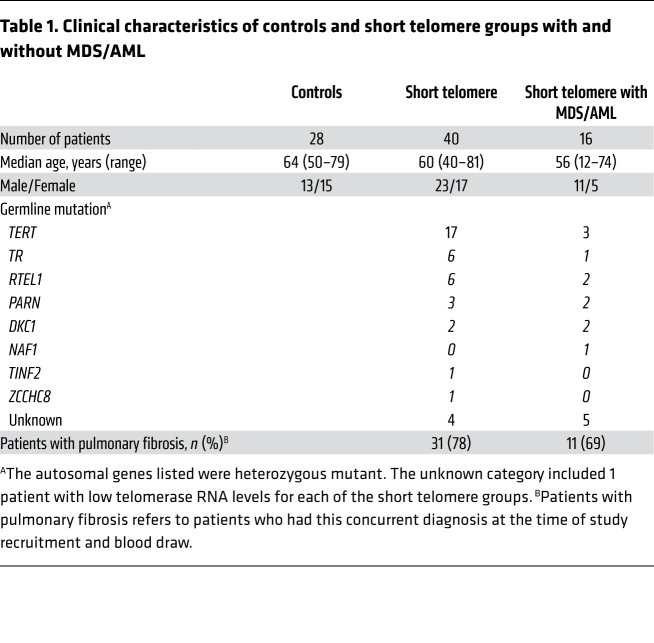
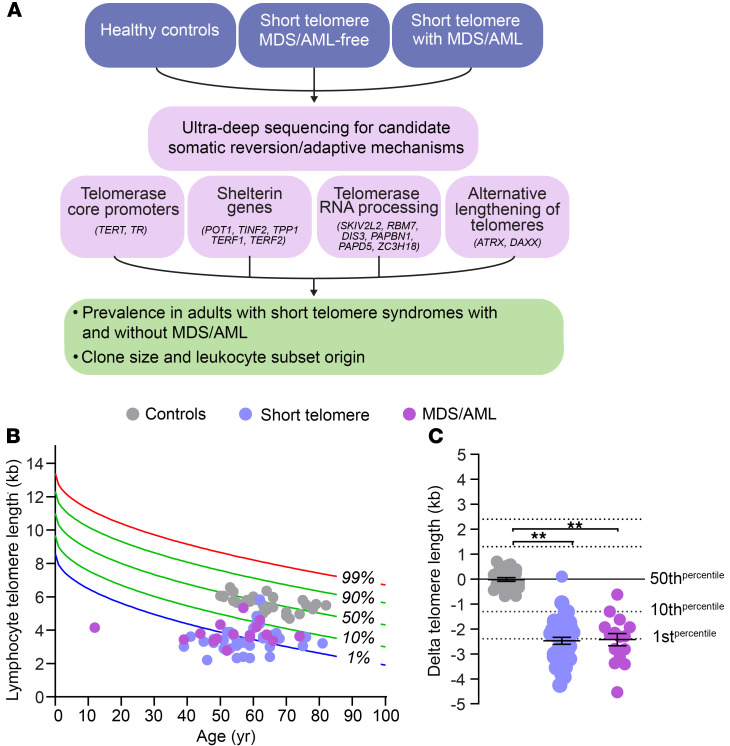
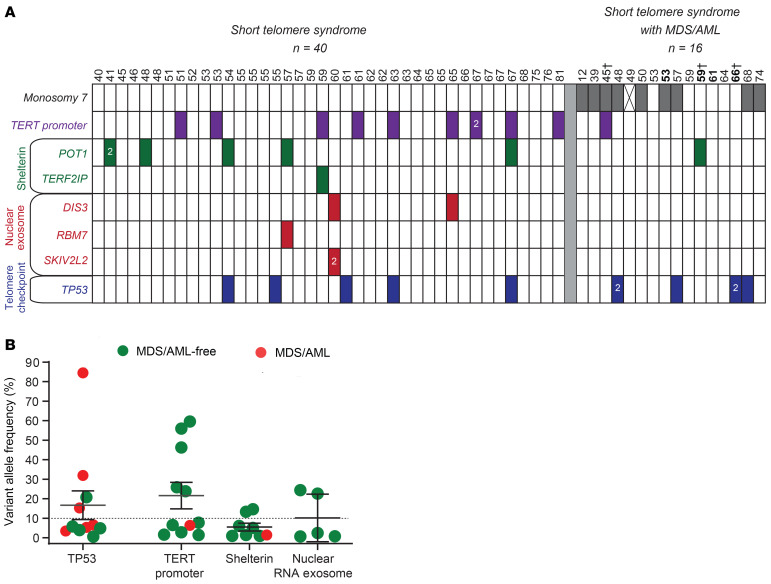
To understand the mechanisms of telomere maintenance in patients with short telomere MDS/AML, we compared the prevalence of somatic mutations among 84 individuals who were divided across 3 groups: healthy controls (mean age 64 years, n = 28), patients with short telomere syndromes who did not have MDS/AML (mean age 60 years, n = 40), and patients with short telomere syndromes who fulfilled World Health Organization criteria for MDS/AML (mean age 56 years, n = 16). Table 1 summarizes their clinical characteristics. Both the control group and patients with short telomere syndromes were selected based on age over 40 years, while all the patients with short telomere MDS/AML with available DNA were included in the analysis. Healthy controls had normal telomere length, while the other 2 groups had comparable abnormalities in telomere length (Figure 1, B and C). We used a customized ultra-deep, error-corrected sequencing platform (13, 14), and designed our experiment to detect low frequency somatic mutations down to 0.5% variant allele frequency (VAF). Using this pipeline, we targeted 16 candidate genes in which mutations could functionally promote telomere maintenance. Their functions were grouped into 4 categories: (a) promoters of the core telomerase components, telomerase reverse transcriptase and telomerase RNA, TERT, and TR, (b) shelterin genes, of which several are mutated in cancer-prone families hypothesized to have long telomere syndromes (15, 16), (c) nuclear RNA exosome genes involved in TR processing and where loss-of-function has been shown to increase mature TR levels, and (d) ATRX and DAXX, known to be mutated in cancers that rely on alternative lengthening of telomeres (ALT) mechanisms (Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI147598DS1). In addition, we examined TP53, since its loss may allow bypass of the short telomere checkpoint (9). Among the 3 groups, we identified 36 clonal somatic mutations: 34 single nucleotide variants (SNVs) and 2 insertions/deletions. They were found in 7 of the 17 candidate genes (35%): TP53, the TERT promoter, POT1, TERF2IP, RBM7, SKIV2L2, and DIS3 (Figure 2, A–D and Table 2). Only 1 was found in a healthy control, and that mutation was a low allele frequency TP53 (VAF 1%) subclinical clonal hematopoiesis mutation. Another 10 TP53 mutations were identified and fell predominantly into the DNA binding domain, and all are recurrently mutated in cancer (Figure 2A). The latter mutations were divided equally among patients with short telomere syndromes with and without MDS/AML (although both patients who carried 2 TP53 mutations had MDS/AML), and the TP53 allele burden was also higher than that found in the patients who were MDS/AML-free (Figure 2A and Figure 3, A and B).
Table 1. Clinical characteristics of controls and short telomere groups with and without MDS/AML.
Figure 1. Study design and telomere lengths.
(A) Flow diagram outlines study design, with gene categories and candidate genes examined by ultra-deep sequencing. (B) Telogram showing lymphocyte telomere length by flow cytometry and FISH relative to age-adjusted nomogram, with each dot representing a patient in 1 of the 3 groups represented in the key. (C) Dot plot shows the difference of each individual’s lymphocyte telomere length relative to the age-adjusted median in B. Error bars represent standard error of the mean. **P < 0.001 (Mann Whitney U test).
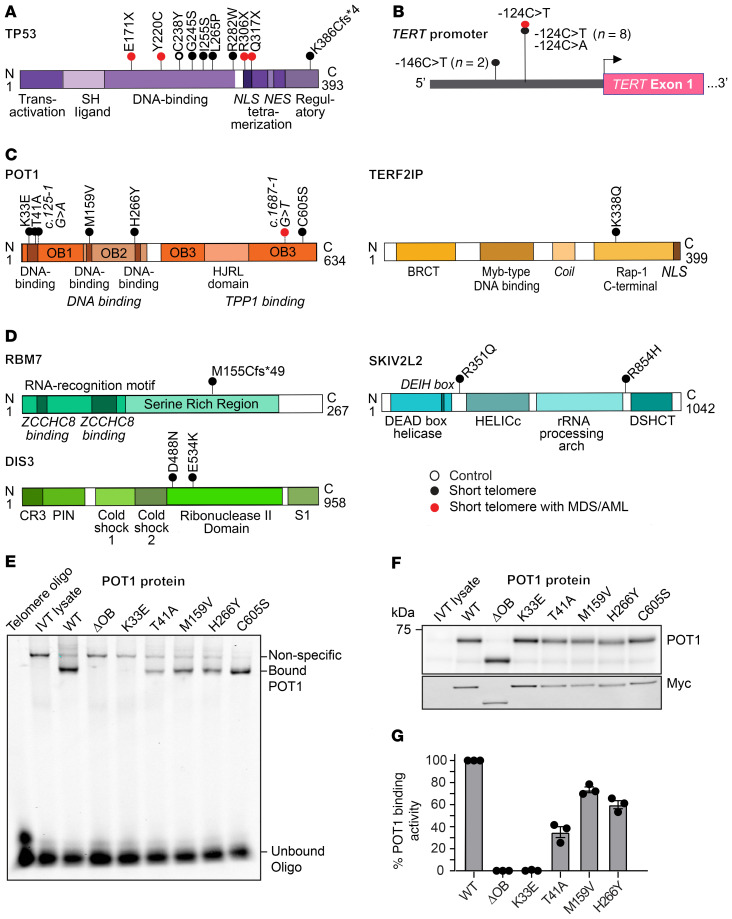
Figure 2. Somatic mutations relative to protein functional domains and POT1 functional studies.
Schema showing somatic mutations relative to mutant protein/gene with conserved domains drawn for (A) TP53, (B) the TERT promoter, (C) shelterin components, and (D) nuclear RNA exosome components, respectively. The key denotes the group in which the mutation was identified, with open circle referring to controls, black circles to patients with short telomere syndromes without MDS/AML, and red circles to patients with short telomere syndromes with MDS/AML. (E) Electrophoretic mobility shift assay (EMSA) for mutant POT1 examining binding capacity to a telomere olignoculeotide. POT1ΔOB refers to a protein deleted for the first DNA binding domain (aa127-635). (F) Immunoblot of in vitro translated products shows stable missense mutant POT1 at expressed levels, with the Myc immunoblot confirming specificity. (G) Mean intensity of binding relative to WT, with error bars representing SEM. The data shown are from 3 EMSA experiments derived from 2 independent in vitro translation reactions.
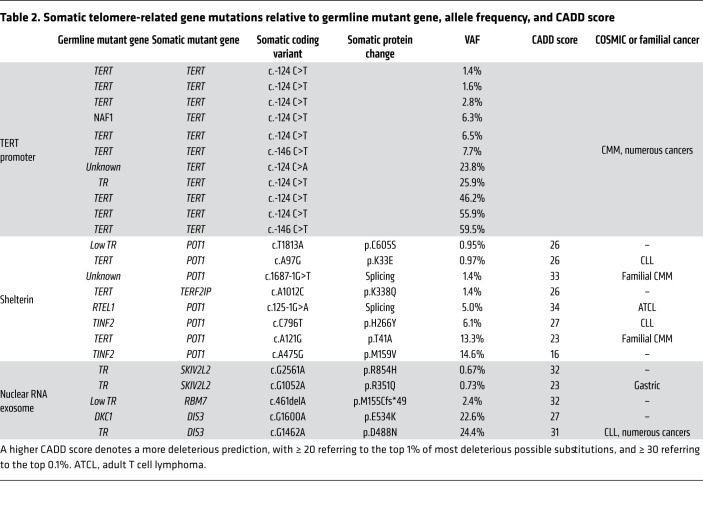
Table 2. Somatic telomere-related gene mutations relative to germline mutant gene, allele frequency, and CADD score.
Figure 3. Prevalence of mutations relative to age and MDS/AML diagnoses.
(A) Chart showing age above and mutant gene identified in each column. Monosomy 7 refers to patients who had any chromosome 7q abnormality including -7, del(7q), or der(1;7) (q10;p10) as determined by karyotype. For patient indicated with X, karyotype information was not available. Ages in bold refer to individuals for whom DNA was derived from a bone marrow aspirate. †Patients who were analyzed for only the 3 most commonly mutant genes on a clinical panel; all others were sequenced on the customized panel. (B) Clone size is plotted by mutant gene or gene group as defined in the labels on the left in A.
All the remaining somatic telomere gene mutations were exclusively in patients with short telomere syndromes with 30% carrying at least 1 mutation (16 of 56 short telomere vs. 0 of 28 controls, P < 0.001, Fisher’s exact test) (Table 2). The most common mutations were in the TERT promoter. They were seen at the –146 and –124 hotspots upstream of TERT’s transcription start site (Figure 2B). Nineteen percent (11 of 56) of the individuals in the combined short telomere groups carried at least one of these mutations. This rate is higher than the 5% to 7% prevalence previously reported in patients with pulmonary fibrosis and dyskeratosis congenita, which used less sensitive sequencing methods (17, 18). These TERT promoter mutations are identical to somatically mutated nucleotides found in numerous cancers, which create de novo ETS-binding sites that upregulate transcription of the intact TERT allele in trans with the germline mutation (17, 19–21). In the 1 patient in whom both canonical sites were mutated, the SNVs were on distinct alleles, indicating they arose in 2 different clones.
The next most common class of mutations was found in POT1, a negative regulator of telomere elongation. Two mutations were found in canonical splice junction sequences (Figure 2C and Table 2). For POT1’s and the missense mutations in the other 4 protein-coding genes, all but one had combined annotation-dependent depletion (CADD) scores greater than or equal to 20, signifying they represent the top 1% of damaging mutations, and 5 had a score of greater than or equal to 30, indicating they were in the top 0.1% of damaging mutations (Table 2). Several of the shelterin gene mutations and mutations in nuclear RNA exosome genes have been documented to be also somatically mutated in cutaneous malignant melanoma (CMM) and chronic lymphocytic leukemia (CLL), among other cancers (Table 2). Somatic mutations at the POT1 p.H266 residue, for example, are the most common telomere maintenance mutation in CLL (22, 23), and 2 mutations, POT1 p.T41A and c.1687-1G>T, have been identified in familial melanoma, a long telomere syndrome phenotype(refs. 15, 24 and our unpublished observations). A third group of mutations was found in RNA processing genes, and the impact of loss-of-function of nuclear RNA exosome targeting (NEXT) complex genes RBM7 and SKIV2L2 and the nuclear RNA exosome essential nuclease DIS3 on increasing TR levels has been well documented (25, 26). We tested the functional consequences of the POT1 missense mutations in an electrophoretic mobility shift assay (EMSA). All 5 missense mutations in the OB folds fell within 3.5 Angstroms of telomeric DNA in the POT1-telomere crystal structure (23, 27) and showed impaired DNA binding to varying degrees (Figure 2, E–G). Notably, the p.K33E somatic mutation phenocopied a mutant that was deleted for the entire first OB fold of POT1 (Figure 2, E–G). In contrast, POT1 p.C605S, which is found in the TPP1-interacting domain (Figure 2C), showed no DNA binding defects. These collective data supported that multiple de novo somatic mutations arise relatively commonly in telomerase and telomere gene mutation carriers. They are functionally impactful and overlap with somatic telomere maintenance mutations seen in cancer.
We next examined if the telomere-related somatic mutations clustered in the patients with MDS/AML, but found a surprising and paradoxical result. Adults with short telomere syndromes who were MDS/AML-free had more mutations than those patients with MDS/AML (RR 4.4, 95% CI 1.2–16.7, P = 0.02, Fisher’s exact test). Among adults without MDS/AML, 35% had at least 1 and 18% had at least 2 somatic mutations (14 and 7 of 40, respectively, Figure 3A). But for patients with MDS/AML, only 13% (2 of 16) carried 1 and none carried 2 mutations. The somatic mutant clones were also on average 3.8-fold larger in MDS/AML-free adults (mean 14.7% vs. 3.9%). Notably, none of the MDS/AML patients had a clonal somatic mutation in a telomere gene with VAF greater than 10% (P = 0.048, Fisher’s exact test; Figure 3B), suggesting this 10% threshold may be a useful predictive threshold if validated in other studies. We assessed the incidence of MDS/AML in the patients who carried somatic reversion mutations and all of them remained MDS/AML-free with a median follow-up of 2.0 years (range 3 months–11 years).
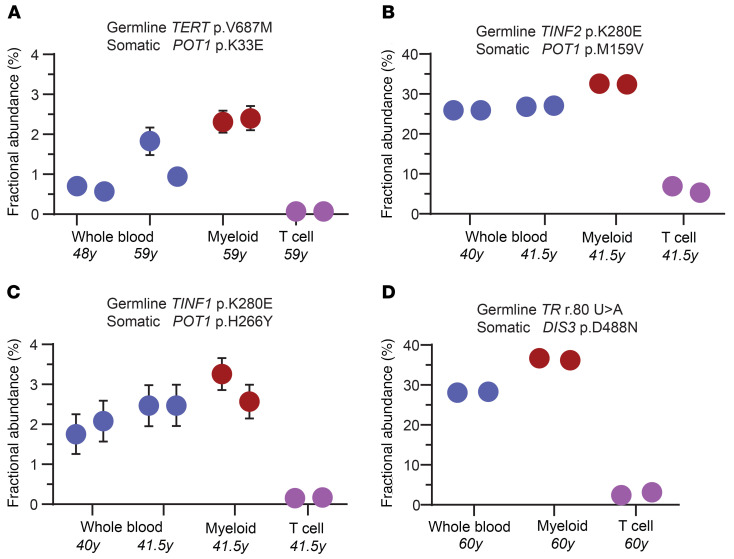
If these somatic mutations correlate negatively with MDS/AML onset, we would predict they would be limited to or enriched in the myeloid lineage. To test this directly, we used droplet digital PCR to quantify the mutant fraction in whole blood and compared it with myeloid and T cell fractions. We studied 4 mutations (POT1 and DIS3) from 3 patients and found they were all enriched in the myeloid lineage while being low or undetectable in the T cell fraction (Figure 4, A–C, POT1, and Figure 4D, DIS3). To definitively test if the somatic mutations we identified were indeed reversion events, we correlated the germline and somatic mutant pathways and found remarkable associations. While TERT promoter mutations predominated in germline TERT mutation carriers (8 of 11), POT1 and TERF2IP mutations were associated with a more heterogeneous spectrum of germline mutations including TERT, TINF2, RTEL1, and PARN (Table 2). This observation is consistent with POT1 loss-of-function mutations playing a role in enhancing telomerase access or processivity (15). By contrast, mutations in the nuclear RNA exosome genes were exclusively detected in patients with germline defects in TR abundance or function. All 3 patients who carried these somatic mutations carried germline mutations in TR itself, the ribonucleoprotein DKC1, or had abnormally low TR levels (Table 2). These data support a model where myeloid-specific adaptation that arises in the highly replicative environment of hematopoiesis, and under the selective pressures of a germline short telomere defect, is associated with an MDS/AML-free state.
Figure 4. Fractional abundance of mutant clones in leukocyte subsets as quantified by ddPCR.
(A–D) ddPCR abundance of mutations is shown at study enrollment and, for 3 of the mutations, at follow-up. The germline and somatic mutations are denoted above and the age at the time of draw is annotated below. DNA for ddPCR analysis was derived from whole blood, the myeloid fraction (CD33+CD66+), and CD3+ T cell fraction, respectively. Each dot refers to the fractional abundance with error bars representing the upper and lower boundaries of the 95% confidence interval of the Poisson distribution. Each reaction was performed twice. Where not visible, the error bars fall within the boundaries of the graphic dot.
Discussion
In the high turnover environment of hematopoiesis, selective pressures imposed by inherited mutations allow advantageous clonal mutations to persist (28). Nearly all previously described somatic reversion mechanisms are direct in that they repair the germline mutant gene itself such as by correcting a frameshift with an insertion or through repair of the mutation by homologous recombination with the WT allele (28–30). Here, we identified several indirect somatic reversion events in the short telomere syndromes, and found they overlap with known telomere maintenance mechanisms in cancer. Their high specificity, such as is the case for the nuclear exosome components in TR mutation carriers, underscores a remarkable versatility of the stressed hematopoietic system to acquire de novo adaptations. We did not observe gross measurable improvements in blood counts or telomere lengthening effects (Supplemental Figure 1A). Nonetheless, the recurrent mutations in the same genes highlight the competitive advantage they provide under the selective pressures of the short telomere environment. Their myeloid enrichment and persistence over 11 years in one patient we studied confirms these mutations arise in a likely primitive myeloid progenitor. The finding of POT1 recurrent mutations across multiple germline defects in TERT and other mutation carriers support a model where POT1 loss-of-function facilitates telomere elongation by improving telomerase access and/or processivity. Notably, several of the somatic mutations we identified are identical to germline mutations seen in cancer-prone families with melanoma (Table 2). Knock-in mutations of some POT1 cancer-associated somatic mutations were recently shown to lengthen telomeres in vitro (31). These data support a model wherein POT1 mutations do not promote genome instability but favor a state of telomere maintenance and lengthening (15, 16).
To our surprise, the adaptive mechanisms we identified, including mutations in the TERT promoter, had a higher prevalence in patients who did not have MDS/AML. These data shed light into how the short telomere bone marrow failure state may evolve to myeloid malignancy. One model is that short telomere progenitors undergoing crisis undergo a fate-determining event that either leads to a functional reversion or a maladaptation such as with loss of chromosome 7. In nearly all the patients we studied, these events were mutually exclusive. Comparing the 35% prevalence of somatic mutations we identified in adult short telomere patients without MDS/AML (Figure 3A), relative to the 10% overall incidence of MDS/AML (6), somatic reversion appears to be a more common and preferred adaptive mechanism. It is also possible that somatic reversion mutations may similarly be a favorable predictor with respect to MDS/AML risk in other inherited bone marrow failure states.
Our data raise the possibility that deep sequencing for at least some of the common mutations we identified may be a useful biomarker for assessing MDS/AML risk in some settings. Idiopathic pulmonary fibrosis is the most common indication of lung transplantation worldwide and it is estimated that 35% of familial pulmonary fibrosis patients carry a germline defect in a telomere maintenance gene (32, 33). In the setting of lung transplantation, short telomere patients are exposed to cytotoxic medications that add selective pressures on hematopoiesis, and the presence of sizable clonal reversion mutations may provide a protective advantage. The median follow-up for patients with telomere-related somatic reversion mutations in our study was 24 months (mean 36 months, range 3–130 months). Additional studies and longer follow-up will be needed to test the predictive utility of somatic reversion testing in clinical settings.
Methods
Patients.
Patients were recruited from 2003 to 2020 as part of the Johns Hopkins Telomere Syndrome Study (6, 34). They were enrolled if they carried a pathogenic mutation in a telomere-related gene and had a personal/family history of a short telomere phenotype, had classic familial short telomere syndromes as defined (7), or had sporadic disease with low telomerase RNA levels (35). The MDS/AML diagnosis was assigned using WHO classification (36). Healthy controls were recruited as previously (37), and were selected for having normal telomere lengths. Cytogenetic abnormalities for the patients with MDS/AML were inferred by karyotype (6). Race data were not collected for this study. Follow-up was last assessed on April 25, 2021.
Germline sequencing, functional studies, and telomere length measurement.
Germline DNA sequencing was performed on peripheral blood DNA using whole genome, exome, or targeted analyses (25, 30, 37). Germline mutations not reported in Alder et al. (37) or Schratz et al. (6) are listed in Supplemental Table 2. Telomerase RNA quantification and DKC1 functional analyses have been detailed previously (25, 38). Peripheral blood telomere length was measured by flow cytometry and fluorescence in situ hybridization (flowFISH) as described (37, 39). All previously unreported germline mutations were deposited in the Telomerase Database (www.telomerase.asu.edu) (40).
Capture design, library preparation, and sequencing.
DNA for somatic analyses was derived from peripheral blood for all 84 individuals in this study, except for 4 individuals from whom it was isolated from a bone marrow aspirate (indicated in Figure 3A). We designed 150 bp read length probes (SureDesign) targeting 37 kb across 17 genes including 2 promoters (Supplemental Table 2). Libraries were prepared following the manufacturer’s instructions (Agilent HaloPlexHS for 1–500 kb target region). Briefly, 400 ng genomic DNA diluted to 14.4 ng/μL were digested in 8 restriction enzyme reactions and hybridized to the custom biotinylated HaloPlexHS probe library incorporating sample indexes and a degenerate 10-nucleotide unique molecular barcode (i.e., UMI, to identify each individual captured DNA molecule) to facilitate base calling accuracy and quantification. Targets were ligated to form circularized fragments, captured using streptavidin beads, and polymerase chain reaction amplified (24 cycles). Target libraries were validated using the Fragment Analyzer (Advanced Analytical Technologies) followed by quantification of fragment smear analysis of 190–640 bp range. All samples were subsequently pooled at a 4 nM concentration. A final AMPure XP magnetic bead cleanup was performed on the pooled library to eliminate any adapter-dimer traces.
HaloPlex library sequencing.
Sequencing was performed on an Illumina HiSeq 2500 using a 150 bp paired-end protocol, and data were demultiplexed following standard protocols. Reads were trimmed and aligned to the reference hg19 genome using the Burrows-Wheeler aligner BWA-MEM in the Agilent Alissa Align and Call v.1.1.2.2 platform. Of 86 samples designed for HaloPlex platform sequencing, 81 eventually fulfilled quality control criteria (2 failed at library preparation, 3 had low coverage). The mean depth of coverage was 12,522× (± 4591 SD) with a median of 10,952× (± 5959 SD). The coverage depth for the TERT promoter was relatively lower and was calculated separately using Samtools (version 1.10, using htslib 1.10). Within the targeted interval (chr5:1295085-1295385, hg19), its coverage was limited to ch5:1295085-1295115 and chr5:1295224-1295385, and these intervals had a mean coverage of 2457× (± 1204 SD) with a median 2421×. Supplemental Table 3 lists the mutations with the number of WT and mutant reads shown.
Three additional patients with MDS/AML were consecutively recruited after the HaloPlex sequencing protocol started, and their DNA was sequenced using a clinically validated dual-indexed targeted method that applied a background error rate analysis to allow robust detection of somatic mutations with allele frequency of at least 1% (6, 41). This pipeline included the 3 most commonly mutated genes in the HaloPlex analysis — TP53, TERT promoter, and POT1 — as well as TINF2. Supplemental Figure 1B lists the mutation types at the base pair level and shows 58% of SNVs were cytosine to thymidine transitions.
Variant calling and analysis.
Variant calling was performed using Agilent Alissa Align and Call v1.1.2.2 software and annotated using ANNOVAR v4162018. To utilize the HaloPlex error-corrected method of analysis, we first collapsed reads originating from the same sample molecule into read families, and duplicates were removed to create consensus reads. Variants were filtered to include only those present in at least 3 unique read families. To exclude sequencing artifacts, we used a quality score of greater than 90 and filtered for a variant score threshold of greater than 0.3 in Alissa. Variants that were present in more than 5% of controls were additionally excluded as alignment artifacts. Variants with allele frequencies of 40% to 60% or greater than 95% were deemed germline and excluded. We finally filtered for variants with minor allele frequencies greater than 0.0005 in gnomAD v.2.1.1 (any VAF), and these were excluded as they were thought to be possibly germline (performed November 1, 2020). The TERT promoter was manually analyzed and curated separately to include variants present in all reads of one or more consensus families at the canonical c.-124 and c.-146 positions, irrespective of variant allele frequency. All of the variants reported were manually verified in Integrative Genomics Viewer. Somatic variants that passed filtering and manual review were submitted to the COSMIC database (July 6, 2021, COSP49927).
Variant interpretation.
Protein schema were drawn based on the longest isoform curated in the Gene Database (National Center for Biotechnology Information, accessed October 21, 2020). Linear protein structures were visualized in SnapGene (v5.1.6), and conserved domains constructed were based on Uniprot (release 2020_05) and the literature. Variants were analyzed for their presence in the COSMIC database (v.92) and CADD score (v1.6) (42).
POT1 gel shift binding assay.
To test the functional significance of POT1 missense mutations, we cloned Myc-FLAG tagged POT1 into T7 expression vector (pcDNA 5/FRT, Addgene) and performed site-directed mutagenesis. A control with POT1ΔOB was generated by deleting the first OB fold (amino acids 127–635). T7 expression vectors were used in an in vitro translation reaction using the TNT Quick Coupled Transcription/Translation System (Promega), according to the manufacturer’s instructions. Briefly, a 50 μL reaction containing 1 μg of expression vector, 20 μM methionine, and 40 μL TNT Quick Master mix containing rabbit reticulocyte lysate was incubated at 30°C for 90 minutes and used immediately for the gel shift assay.
The POT1 binding assays were performed using the Odyssey Infrared EMSA Kit (LI-COR). Briefly, a 20 μL reaction mixture was prepared containing 2 μL of the IVT product, 10 nM IRDye 800-labeled, single-stranded telomeric oligonucleotide, 2.5 μM nonspecific ssDNA, and 1 μg poly(dI-dC) in binding buffer containing 100 mM Tris, 500 mM KCl, 12.5 mM DTT, 10 mM EDTA, and 0.25% Tween 20. We used the telomere oligonucleotide 5′GGTTAGGGTTAGGGTTAGGG and the nonspecific ssDNA oligonucleotide 5′TTAATTAACCCGGGGATCCGGCTTGATCAACGAATGATCC as per Baumann et al. (43). Reactions were incubated for 30 minutes at room temperature and protein-DNA complexes were resolved by gel electrophoresis on a 10% polyacrylamide Tris-Borate EDTA at 100V for 2 hours at 4°C. Gels were visualized using the Li-Cor Odyssey Imaging System. Quantification was done using ImageJ (2.1.0).
Western blot.
Using the NuPAGE SDS-PAGE gel system, 1 μL of in vitro translation reaction was run on 8% Bis-Tris gels with MOPS-SDS running buffer at 150V and transferred to a PVDF membrane using the iBlot 2 Dry Blotting System on setting P0 (Thermo Fisher Scientific). Membranes were blocked for 1 hour before antibody staining. Membranes were stained first with anti-POT1 antibody (rabbit, NB500-176, 1:500, Novus) and subsequently with anti-rabbit HRP antibody (goat, 1:20,000, Cell Signaling Technology) before visualization via chemiluminescence (SuperSignal West Pico Plus Chemiluminescent substrate kit) using ImageQuant. Membranes were then stripped for 15 minutes, using New Blot IR Stripping buffer, and blocked again for 1 hour before staining with anti-Myc antibody (mouse, clone 4A6, 1:1000; Millipore). Membranes were subsequently stained with anti-mouse IRDye secondary antibody as appropriate (IR680, donkey, 1:10,000, LI-COR) before visualization on an Odyssey scanner (LI-COR). Quantification was done using ImageJ (2.1.0).
Cell fractionation.
Aliquots of fresh blood or frozen Ficoll-separated peripheral blood cells were fractionated using the EasySep Human Myeloid Positive Selection and EasySep Human T Cell Enrichment kits (StemCell Technologies). DNA was extracted from the fractionated subsets using the MagMAX DNA Multi-Sample Ultra 2.0 Kit (Thermo Fisher Scientific) and quantified using the Quibit dsDNA HS Assay kit (Thermo Fisher Scientific).
Droplet digital PCR.
Droplet digital PCR (ddPCR) was performed according to the manufacturer’s specifications on a QX200 system (Bio-Rad). Sequence-specific primers and probes were designed using the Bio-Rad digital assay design engine: POT1 p.K33E (dHsaMDS726207399), POT1 p.M159V (dHsaMDS784505022), POT1 p.H266Y (dHsaMDS401027863), and DIS3 p.D488N (HsaMDS2512362). An annealing temperature of 58°C was used except for the POT1 p.K33E (54°C) and DIS3 p.D488N (54°C). The WT and mutant probes were conjugated with HEX and FAM reporters, respectively. PCR amplicons were sequence-verified by cloning the products (TOPO TA Cloning Kit, Invitrogen). For each target, the mean number of droplets analyzed was 11,890 ± 2081 SD and all reactions had more than 9000 events analyzed. Data were analyzed in QX Manager (version 1.2-STD, Bio-Rad).
Statistics.
Graphs were generated and statistical calculations were performed using GraphPad Prism 8.0. The respective statistical tests are included in the text and figure legends and a P value less than 0.05 was considered significant. All P values shown are 2-sided.
Study approval.
The research was approved by Johns Hopkins Medicine’s Institutional Review Board, and patients gave written informed consent.
Author contributions
KES designed the study and analyzed the clinical and genetic data. KES and VG selected the candidate genes. VG prepared the libraries with LKS. ZLC performed the POT1 functional studies and analyzed ddPCR data. EAD performed computational analyses. ZX performed the fractionation and ddPCR experiments. LF supported the sequencing analysis pipeline design. PDS analyzed clinical data. MA oversaw the project and wrote the paper with KES. All the authors reviewed and approved the manuscript.
Supplementary Material
Acknowledgments
We would like to acknowledge helpful conversations with Dustin Gable, and technical support from the Johns Hopkins Genomics team, the Johns Hopkins Genetics Resources Core Facility, and the Bloomberg School of Public Health Flow Cytometry Core. We are grateful for technical support from the Agilent team of Josh Zhiyong Wang, Gregory Miles, and Weiwei Liu. This work was supported by R01CA225027 and R01HL119476, the Commonwealth Foundation, the S&R Foundation Kuno Award, and the Williams Foundation (to MA); T32HL007525, a grant from the Vera and Joseph Dresner Foundation, the MacMillan Pathway to Independence Award, and the American Society of Hematology (ASH) Scholar Award (to KES); F32HL142207 (to VG); the Johns Hopkins Research Program for Medical Students (to ZLC); T32 GM136577 (to EAD); and the Turock Scholars Fund to the Telomere Center at Johns Hopkins (to KES and EAD). The authors would also like to acknowledge a gift in the name of P. Godrej.
Version 1. 08/03/2021
In-Press Preview
Version 2. 09/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(18):e147598.https://doi.org/10.1172/JCI147598.
Contributor Information
Kristen E. Schratz, Email: kschrat1@jhmi.edu.
Valeriya Gaysinskaya, Email: vgaysin1@alumni.jh.edu.
Zoe L. Cosner, Email: zcosner1@jhmi.edu.
Emily A. DeBoy, Email: edeboy@jhmi.edu.
Zhimin Xiang, Email: zxiang2@jhmi.edu.
Laura Kasch-Semenza, Email: lkasch@jhmi.edu.
Liliana Florea, Email: florea@jhu.edu.
Pali D. Shah, Email: pdedhiy2@jhmi.edu.
Mary Armanios, Email: marmani1@jhmi.edu.
References
- 1.Keel SB, et al. Genetic features of myelodysplastic syndrome and aplastic anemia in pediatric and young adult patients. Haematologica. 2016;101(11):1343–1350. doi: 10.3324/haematol.2016.149476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calado RT, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106(4):1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reilly CR, et al. The clinical and functional effects of TERT variants in myelodysplastic syndrome. Blood. 2021;138(10):898–911. doi: 10.1182/blood.2021011075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluteau O, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131(7):717–732. doi: 10.1182/blood-2017-09-806489. [DOI] [PubMed] [Google Scholar]
- 5.Schratz KE, Armanios M. Cancer and myeloid clonal evolution in the short telomere syndromes. Curr Opin Genet Dev. 2020;60:112–118. doi: 10.1016/j.gde.2020.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schratz KE, et al. Cancer spectrum and outcomes in the Mendelian short telomere syndromes. Blood. 2020;May 28 135(22):1946–1956. doi: 10.1182/blood.2019003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parry EM, et al. Syndrome complex of bone marrow failure and pulmonary fibrosis predicts germline defects in telomerase. Blood. 2011;117(21):5607–5611. doi: 10.1182/blood-2010-11-322149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merck SJ, Armanios M. Shall we call them “telomere-mediated”? Renaming the idiopathic after the cause is found. Eur Respir J. 2016;48(6):1556–1558. doi: 10.1183/13993003.02115-2016. [DOI] [PubMed] [Google Scholar]
- 9.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97(4):527–538. doi: 10.1016/S0092-8674(00)80762-X. [DOI] [PubMed] [Google Scholar]
- 10.Hemann MT, et al. Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol Biol Cell. 2001;12(7):2023–2030. doi: 10.1091/mbc.12.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alder JK, et al. Telomere dysfunction causes alveolar stem cell failure. Proc Natl Acad Sci U S A. 2015;112(16):5099–5104. doi: 10.1073/pnas.1504780112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13(10):693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia J, et al. Somatic mutations and clonal hematopoiesis in congenital neutropenia. Blood. 2018;131(4):408–416. doi: 10.1182/blood-2017-08-801985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncavage EJ, et al. Mutation clearance after transplantation for myelodysplastic syndrome. N Engl J Med. 2018;379(11):1028–1041. doi: 10.1056/NEJMoa1804714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNally EJ, et al. Long telomeres and cancer risk: the price of cellular immortality. J Clin Invest. 2019;129(9):3474–3481. doi: 10.1172/JCI120851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley SE, Armanios M. The short and long telomere syndromes: paired paradigms for molecular medicine. Curr Opin Genet Dev. 2015;33:1–9. doi: 10.1016/j.gde.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maryoung L, et al. Somatic mutations in telomerase promoter counterbalance germline loss-of-function mutations. J Clin Invest. 2017;127(3):982–986. doi: 10.1172/JCI91161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez-Rodrigues F, et al. Pathogenic TERT promoter variants in telomere diseases. Genet Med. 2019;21(7):1594–1602. doi: 10.1038/s41436-018-0385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell RJA, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348(6238):1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 22.Herling CD, et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood. 2016;128(3):395–404. doi: 10.1182/blood-2016-01-691550. [DOI] [PubMed] [Google Scholar]
- 23.Ramsay AJ, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45(5):526–530. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 24.Shen E, et al. POT1 mutation spectrum in tumour types commonly diagnosed among POT1-associated hereditary cancer syndrome families. J Med Genet. 2020;57(10):664–670. doi: 10.1136/jmedgenet-2019-106657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gable DL, et al. ZCCHC8, the nuclear exosome targeting component, is mutated in familial pulmonary fibrosis and is required for telomerase RNA maturation. Genes Dev. 2019;33(19–20):1381–1396. doi: 10.1101/gad.326785.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tseng CK, et al. Human telomerase RNA processing and quality control. Cell Rep. 2015;13(10):2232–2243. doi: 10.1016/j.celrep.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 27.Lei M, et al. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11(12):1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 28.Revy P, et al. Somatic genetic rescue in Mendelian haematopoietic diseases. Nat Rev Genet. 2019;20(10):582–598. doi: 10.1038/s41576-019-0139-x. [DOI] [PubMed] [Google Scholar]
- 29.Jongmans MC, et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am J Hum Genet. 2012;90(3):426–433. doi: 10.1016/j.ajhg.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alder JK, et al. Exome sequencing identifies mutant TINF2 in a family with pulmonary fibrosis. Chest. 2015;147(5):1361–1368. doi: 10.1378/chest.14-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim WT, et al. Cancer-associated POT1 mutations lead to telomere elongation without induction of a DNA damage response. EMBO J. 2021;40(12):e107346. doi: 10.15252/embj.2020107346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silhan LL, et al. Lung transplantation in telomerase mutation carriers with pulmonary fibrosis. Eur Respir J. 2014;44(1):178–187. doi: 10.1183/09031936.00060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley SE, et al. Telomerase mutations in smokers with severe emphysema. J Clin Invest. 2015;125(2):563–570. doi: 10.1172/JCI78554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonassaint NL, et al. The gastrointestinal manifestations of telomere-mediated disease. Aging Cell. 2013;12(2):319–323. doi: 10.1111/acel.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley SE, et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. Sci Transl Med. 2016;8(351):351ra107. doi: 10.1126/scitranslmed.aaf7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sherdlow SH, et al, eds. WHO Classification Of Tumours Of Haematopoietic and Lymphoid Tissues. World Health Organization; 2017. [Google Scholar]
- 37.Alder JK, et al. Diagnostic utility of telomere length testing in a hospital-based setting. Proc Natl Acad Sci U S A. 2018;115(10):E2358–E2365. doi: 10.1073/pnas.1720427115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaysinskaya V, et al. Synonymous mutation in DKC1 causes telomerase RNA insufficiency manifesting as familial pulmonary fibrosis. Chest. 2020;158(6):2449–2457. doi: 10.1016/j.chest.2020.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baerlocher GM, et al. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat Protoc. 2006;1(5):2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 40.Podlevsky JD, et al. The telomerase database. Nucleic Acids Res. 2008;36(database issue):D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng G, et al. The diagnostic utility of targeted gene panel sequencing in discriminating etiologies of cytopenia. Am J Hematol. 2019;94(10):1141–1148. doi: 10.1002/ajh.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baumann P, et al. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol Cell Biol. 2002;22(22):8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.