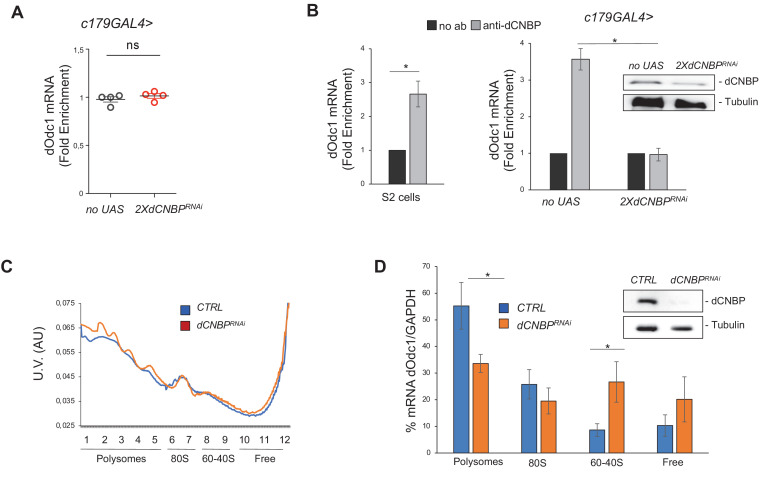
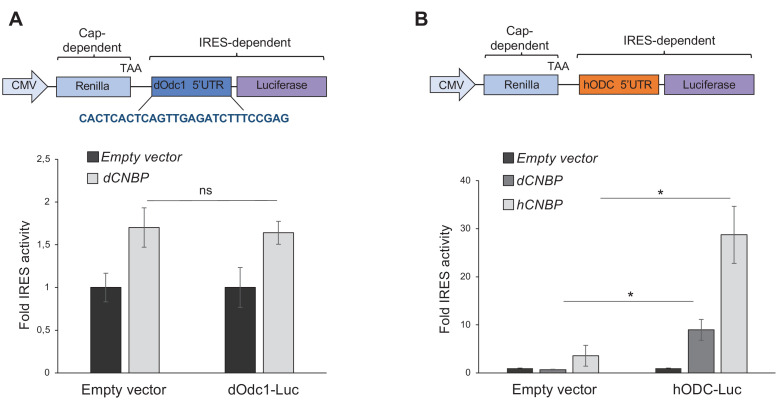
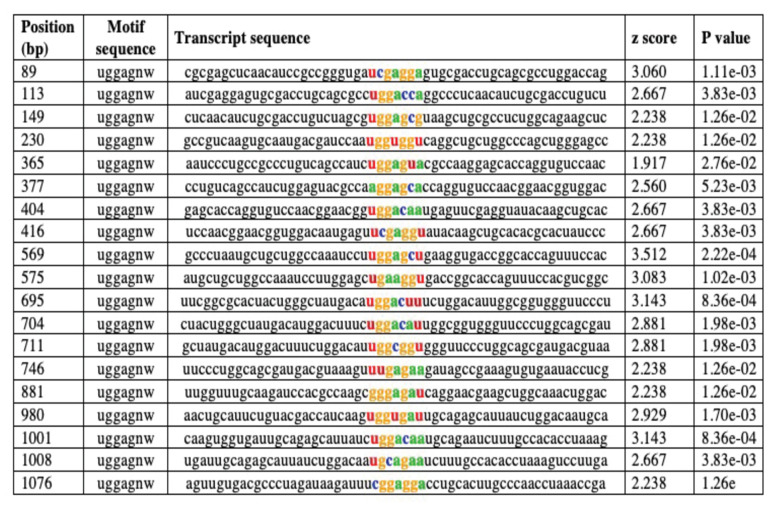
Figure 6. dCNBP controls polyamine metabolism through the binding and the translational control of dOdc mRNA.
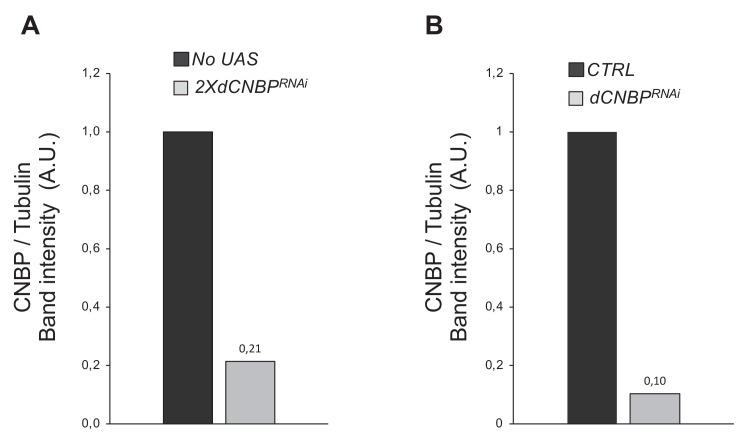
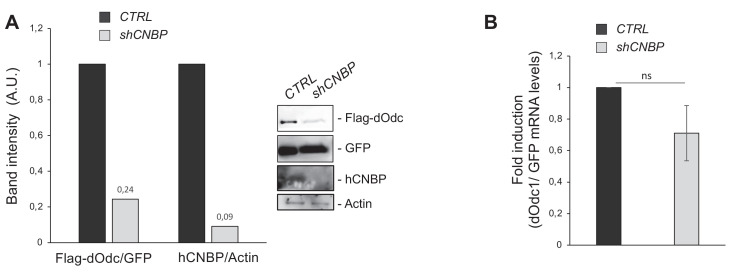
(A) dOdc1 mRNA levels (qPCR), normalized with the housekeeping RPL11 mRNA third instar larvae bearing c179GAL4 driver alone (no UAS) or in combination with UAS-dCNBPRNAi-16283; UAS-dCNBPRNAi-16284 (2XdCNBPRNAi). ns, not significant in unpaired t-test. Dots correspond to four independent biological replicates; bars indicate the mean and SEM. (B) Cellular nucleic acid-binding protein (CNBP) binds dOdc1 mRNA. qRT-PCR analysis on mRNAs immunoprecipitated by anti-dCNBP antibody or control IgG antisera in S2 cells extracts (left graph), or in dCNBP-depleted (2XdCNBPRNAi) or not (no UAS) larval extracts (right graph). The results are indicated as fold difference, relative to IgG. Error bars represent SEM of three independent experiments; *p < 0.05, in t-test. The presence of dCNBP in c179GAL4>2XUASdCNBPRNAi or control (no UAS) larval carcasses was analyzed by western blotting (right). Tubulin, loading control. (C) Representative polysome profiles (of at least three independent experiments) of dCNBP-deficient (dCNBPRNAi) or control (CTRL) S2 cells. Cytoplasmic lysates were fractionated on 15–50% sucrose gradients. (D) qPCR analysis of dOdc1 mRNA loaded in the different polysome fractions, GADPH was used to normalize the values. (*p < 0.05, t-test. Error bars represent SEM of experiments performed in quadruplicates and repeated at least three times.) The presence of dCNBP in interfered or not interfered S2 cells was analyzed by western blotting (right). Tubulin, loading control. All full data in Figure 6—source data 1.