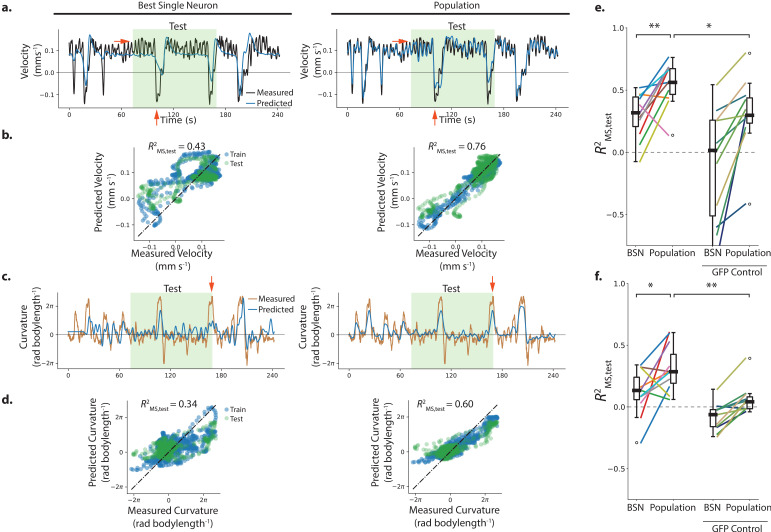
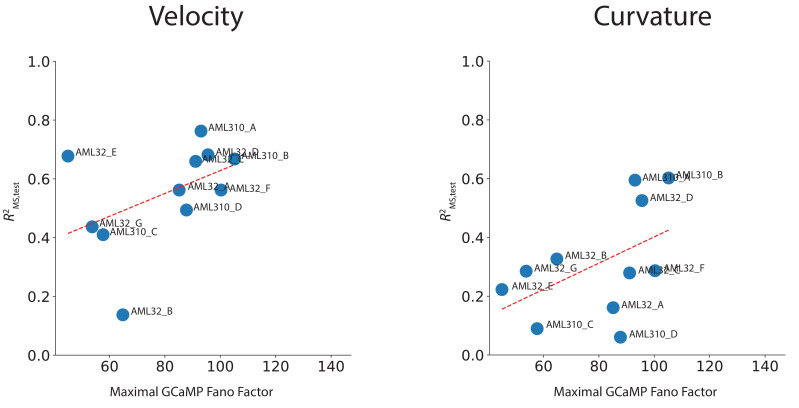
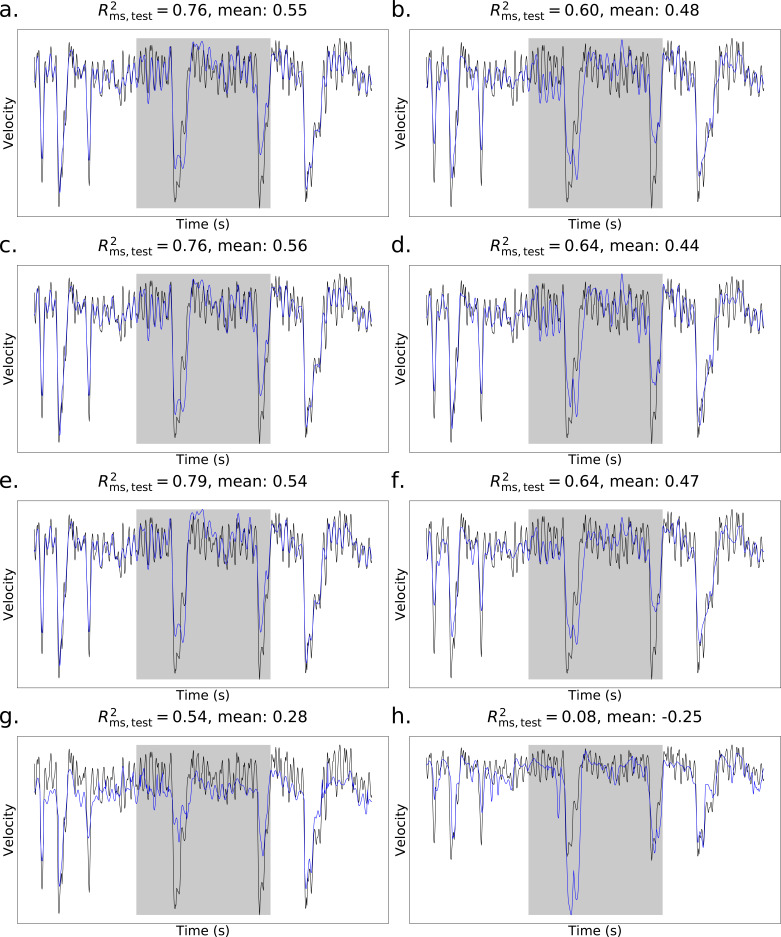
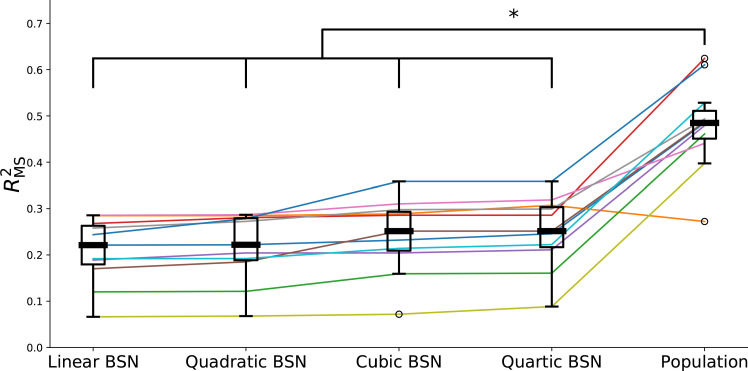
Figure 3. Population neural activity decodes locomotion.
(a–d) Performance of the best single neuron (BSN) is compared to a linear population model in decoding velocity and body curvature for the exemplar recording AML310_A shown in Figure 1. (a) Predictions on the held-out test set are compared to measured velocity. Light green shaded region indicates held-out test set. Red arrows indicate examples of features that the population captures better than the BSN. (b) Performance is reported as a coefficient of determination evaluated on the mean-subtracted held-out test data (green points). (c,d) Model predictions are compared to measured curvature. (e) Performance of velocity decoding is shown for recordings of individuals (strain AML310 and AML32) and for recordings of GFP control animals lacking a calcium indicator (strain AML18). Two-sided Wilcoxon rank test is used to test significance of population performance compared to BSN, . Welch’s unequal variance t-test is used to test significance of population performance compared to GFP control, . (f) Performance of curvature decoding is shown for all recordings. Each recording is colored the same as in e. and for comparisons of population performance to that of BSN, and GFP control, respectively.