Abstract
Background
Understanding the complexities of immune memory to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is key to gain insights into the durability of protective immunity against reinfection.
Objective
We sought to evaluate the immune memory to SARS-CoV-2 in convalescent patients with longer follow-up time.
Methods
SARS-CoV-2–specific humoral and cellular responses were assessed in convalescent patients with coronavirus disease 2019 (COVID-19) at 1 year postinfection.
Results
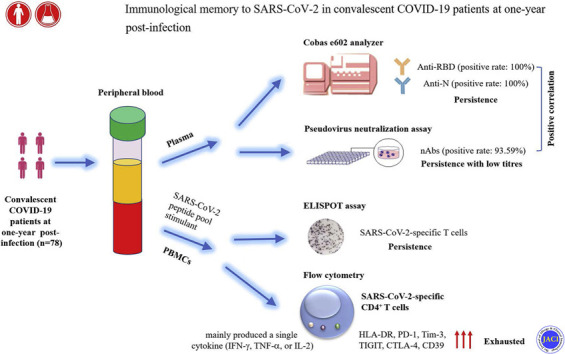
A total of 78 convalescent patients with COVID-19 (26 moderate, 43 severe, and 9 critical) were recruited after 1 year of recovery. The positive rates of both anti–receptor-binding domain and antinucleocapsid antibodies were 100%, whereas we did not observe a statistical difference in antibody levels among different severity groups. Accordingly, the prevalence of neutralizing antibodies (nAbs) reached 93.59% in convalescent patients. Although nAb titers displayed an increasing trend in convalescent patients with increased severity, the difference failed to achieve statistical significance. Notably, there was a significant correlation between nAb titers and anti–receptor-binding domain levels. Interestingly, SARS-CoV-2–specific T cells could be robustly maintained in convalescent patients, and their number was positively correlated with both nAb titers and anti–receptor-binding domain levels. Amplified SARS-CoV-2–specific CD4+ T cells mainly produced a single cytokine, accompanying with increased expression of exhaustion markers including PD-1, Tim-3, TIGIT, CTLA-4, and CD39, while the proportion of multifunctional cells was low.
Conclusions
Robust SARS-CoV-2–specific humoral and cellular responses are maintained in convalescent patients with COVID-19 at 1 year postinfection. However, the dysfunction of SARS-CoV-2–specific CD4+ T cells supports the notion that vaccination is needed in convalescent patients for preventing reinfection.
Key words: SARS-CoV-2, COVID-19, neutralizing antibodies, CD4+ T-cell responses, immune memory
Abbreviations used: COVID-19, Coronavirus disease 2019; M, Membrane; N, Nucleocapsid; nAb, Neutralizing antibody; ORF, Opening reading frame; RBD, Receptor-binding domain; S, Spike; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2
Graphical abstract
Coronavirus disease 2019 (COVID-19), the emerging infectious disease caused by a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is still the greatest threat to public health worldwide.1, 2, 3 Globally, as of June 10, 2021, there have been 174 million confirmed cases of COVID-19, including 3.8 million deaths, reported to the World Health Organization. Currently, the development of vaccines is the most important strategy against COVID-19, depending on further clarification of immune memory in convalescent patients.
The production of SARS-CoV-2–specific antibodies, especially neutralizing antibodies (nAbs), is key for protecting against viral reinfection and provides insight into the design of vaccination strategies.4 , 5 Generally, SARS-CoV-2–specific IgM antibody level peaks at week 3 and then declines, whereas IgG antibodies to spike (S) protein can persist long-term, even beyond 6 months after infection.6, 7, 8, 9, 10 However, nAb titers gradually decline after an initial peak in convalescent patients, and most convalescent plasma samples obtained from individuals who recover from COVID-19 do not contain high levels of neutralizing activity.11 , 12 Notably, nAb titers are positively correlated with COVID-19 severity.13, 14, 15, 16 It is noteworthy that serum IgG antibodies to SARS-CoV-2 receptor-binding domain (RBD) of S protein correlate well with nAb titers, which suggests that commercially available anti-RBD antibodies can serve as useful surrogates for nAb testing.16, 17, 18 Although previous studies have observed persistent humoral responses in convalescent patients, especially in those with severe disease for at least 6 months, how long nAbs will persist or whether they will provide protection from reinfection needs to be further studied.
SARS-CoV-2–specific T-cell responses are central for the control of viral infections and provide immunologic memory that enables long-lasting protection, especially in individuals with negative or low titers of nAbs.19, 20, 21 Emerging data indicate that SARS-CoV-2–specific CD8+ and CD4+ T cells targeting different viral proteins are detectable in up to 70% and 100% of convalescent individuals, respectively.22, 23, 24, 25 More specifically, the membrane (M), S, and nucleocapsid (N) protein each account for 11% to 27% of the total CD4+ T-cell responses, with additional responses commonly targeting nonstructural protein 3, nonstructural protein 4, opening reading frame (ORF)-3a, and ORF-8, providing evidence that diversity of SARS-CoV-2 T-cell responses is common in convalescent patients with COVID-19.22 Notably, there is a strong correlation between the number of SARS-CoV-2–specific T cells and nAb titers.5 , 24 Although a recent study has found that SARS-CoV-2–specific T-cell responses can be detected in convalescent patients at 6 to 7 months postinfection,26 the duration of SARS-CoV-2–specific T-cell memory, including the abundance, phenotype, and functional capacity, still needs to be further elucidated in patients with a longer recovery period.
A deep elucidation of immune memory to SARS-CoV-2 requires evaluation of its core elements, such as nAbs and CD4+ T cells. Understanding the complexities of immune memory to SARS-CoV-2 is key to gain insights into the likelihood of durability of protective immunity against reinfection. In this study, we assessed the SARS-CoV-2–specific anti-RBD and anti-N antibodies, nAbs, and CD4+ T-cell responses simultaneously in convalescent COVID-19 cases, extending up to 1 year after infection. For the first time, our study provided evidence that although robust SARS-CoV-2–specific humoral and cellular responses were maintained in convalescent patients for as long as 1 year, the low titers of nAbs and exhausted function of SARS-CoV-2–specific CD4+ T cells indicated that vaccination was needed in convalescent patients for preventing reinfection.
Methods
Patients
Between March 2021 and April 2021, a total of 78 convalescent patients with COVID-19 were recruited to this study. All convalescent patients had been diagnosed with COVID-19 between January 2020 and March 2020 by positive SARS-CoV-2 real-time RT-PCR test result in Tongji Hospital, Wuhan, China. The enrolled convalescent patients were divided into moderate, severe, and critical groups according to the guideline of diagnosis and treatment of COVID-19 of the National Health Commission of China as follows: (1) moderate, patients have saturation of oxygen greater than or equal to 94% on room air during hospitalization; (2) severe, patients have sign of hypoxia (respiration rate ≥ 30 times/min, saturation of oxygen ≤ 93%, or ratio of arterial partial pressure of oxygen to fraction of inspired oxygen ≤ 300 mm Hg); and (3) critical, patients have respiratory failure requiring mechanical ventilation, septic shock, and/or multiple organ dysfunction. After treatment, all enrolled cases met the discharged criteria and were discharged before March 31, 2020. The demographic, clinical information, and outcome data were collected from electronic medical records. This study was approved by the Ethical Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China (institutional review board ID: TJ-IRB20210137). Written informed consent was obtained from all the participants.
SARS-CoV-2 antibody detection
Blood samples were collected from study participants. SARS-CoV-2 antibodies were measured using the quantitative Elecsys anti-RBD and semi-quantitative Elecsys anti-N (both measuring total immunoglobulin levels) on the Cobas e602 analyzer (Roche Diagnostics, Rotkreuz, Switzerland). Results for the quantitative Elecsys anti-RBD antibodies are reported as concentrations (U/mL), with a manufacturer’s cutoff of more than 0.8 U/mL considered as positive. Results for the Elecsys anti-N antibodies are reported as cutoff index (signal sample/cutoff or signal calibrator), with values more than 1 considered as positive. Quality controls and coefficients of variation for both assays are provided in Table E1 in this article’s Online Repository at www.jacionline.org. The values of anti-RBD antibodies were log10 transformed before analysis.
Neutralization antibody detection using pseudovirus neutralization assay
Pseudovirus neutralization assay was performed as our previous study with minor modifications.14 A full-length codon-optimized s gene of SARS-CoV-2 was first synthesized and cloned into the lentivirus vector GV367 (Genechem, Shanghai, China), and then used to generate an eGFP-coexpressing pseudovirus by cotransfection into HEK293T cells (CRL-11268) with the other 2 viral packaging help vectors pHelper1.0 and pHelper2.0 (Genechem, Shanghai, China). Forty-eight hours after transfection, the supernatants were collected after centrifugation at 4000g for 10 minutes at 4°C, and further filtrated with a 0.45-μm filter. The recombinant pseudovirus was further purified by centrifugation at 25000 rpm for 2 hours at 4°C and diluted with PBS. The titer of recombinant pseudovirus was quantified by fluorometry and RT-quantitative PCR targeting the s gene. The SARS-CoV-2 pseudovirus neutralization assay was carried out on hACE2-COS7 cells (Vitalstar, Beijing, China) in a 96-well plate. Fifty microliter serial 2-fold diluted sera from 1:10 to 1:320 from each serum sample were prepared, and equal volumes of SARS-CoV-2 pseudovirus were added and the plates were preincubated at 37°C for 1 hour. Twenty-four hours before infection, 100 μL of 5 × 103 hACE2-COS7 cells were added into each well of a 96-well plate. After washing and adding 100 μL fresh culture medium, cells were incubated with 100 μL of sera-pseudovirus mixture for 48 hours. The cells were collected with 200 μL of digestion solution and used to determine the number of eGFP-expressing cells by flow cytometry. The positive rate of eGFP-expressing cells was calculated after collecting 1000 cells. Experiments were repeated twice. The neutralization rate (%) for different dilutions was calculated as follows: the titer of neutralization antibody for each serum sample was expressed as the half-maximal neutralizing titer. Half-maximal neutralizing titer of each serum sample was determined as the highest dilution ratio of serum with 50% neutralization rate. The cutoff value of half-maximal neutralizing titer was defined by receiver-operating characteristic curve analysis to discriminate between convalescent patients with COVID-19 and healthy controls (no COVID-19 exposure), with values more than 25 considered as positive.
SARS-CoV-2–specific ELISPOT assay
Heparinized blood samples were collected from study participants, and PBMCs were isolated by using Ficoll-Hypaque density gradients. PBMCs (2.5 × 105) were added to 96-well plates precoated with anti–IFN-γ antibody (Millipore, Temecula, Calif) in 100 μL of AIM-V medium containing 10% FBS, 50 μg/mL streptomycin, and 10 μg/mL gentamicin (ThermoFisher Scientific, Grand Island, NY). SARS-CoV-2 peptide pool (2 μg/mL, MABTECH, Stockholm, Sweden) derived from the S, N, M, ORF-3a, and ORF-7a proteins was used to stimulate cells. PHA (15 μg/mL, Sigma-Aldrich, Dorset, UK) and AIM-V medium were added and used for positive control and negative control well, respectively. Plates were incubated for 16 to 20 hours at 37°C with 5% CO2, washed with PBS, and developed using an anti–IFN-γ antibody conjugate and substrate to detect the presence of secreted IFN-γ. Spot-forming cells were counted with an automated ELISPOT reader (CTL Analyzers, Cleveland, Ohio). Results were considered positive if the spot amounts in the SARS-CoV-2 peptide pool minus the negative control were more than 5 spot-forming cells.
The cytokine secretion capability and phenotype of SARS-CoV-2–specific CD4+ T cells
PBMCs were isolated from heparinized blood samples and stimulated with SARS-CoV-2 peptide pool or medium in the presence of 2 μM monensin (eBioscience, San Diego, Calif) for 24 hours. After culture, the cells were collected for flow cytometry analysis. Fluorescence-labeled mAbs against the following antigens were added to the cell suspensions as follows: fixable viability stain, CD3, CD4, CD45RA, CCR7, HLA-DR, PD-1, Tim-3, TIGIT, CTLA-4, and CD39 (BD Biosciences, San Jose, Calif). All these cell suspensions were incubated for 30 minutes on ice. In some experiments, cells were fixed and permeabilized, and stained with anti–IL-2, anti–IFN-γ, anti–TNF-α, anti–IL-4, and anti–IL-17 mAbs (eBioscience). Isotype controls with irrelevant specificities were included as negative controls. Fixable viability stain was used to exclude dead cells from analysis. After washings, the pellets were resuspended in 300 μL staining buffer, followed by analysis with FACSCanto flow cytometer (BD Biosciences).
Statistical analysis
The results are presented as mean ± SD, or as median with interquartile range when appropriate. Continuous variables were compared with Mann-Whitney U test or 1-way ANOVA test. Fisher exact test was used for categorical data. Receiver-operating characteristic curve analysis was performed to determine the best cutoff value of nAb titers for discriminating between convalescent patients with COVID-19 and healthy controls. Spearman rank correlation test for nonparametric data was used to analyze the relationship between 2 factors. Statistical significance was determined as P < .05 (∗P< .05, ∗∗P < .01, ∗∗∗P < .001). Statistical analyses were performed using SPSS version 19.0 (SPSS, Chicago, Ill), GraphPad Prism 8.0 (San Diego, Calif).
Results
Participant characteristics
A total of 78 convalescent patients with COVID-19, including 26 moderate, 43 severe, and 9 critical cases, were recruited after 1 year of recovery. All recovered patients enrolled in this study had a history of hospitalization in Tongji Hospital between January 2020 and March 2020. The demographic and clinical information of the participants when they were in hospital was collected from electronic medical records and is presented in Table E2 in this article’s Online Repository at www.jacionline.org. No significant difference in age, sex, clinical symptoms, and comorbidities was observed among different groups.
The current status of convalescent patients with COVID-19
The current health status of all enrolled convalescent patients with COVID-19 at 1 year postinfection was assessed by clinicians and the detailed information is presented in Table I . The average days from onset of illness to serum sampling in these recovered patients were 381 days. Most participants demonstrated normal routine laboratory test results including blood routine, liver function, renal function, myocardial function, and inflammatory biomarker IL-6. A few indicators such as serum creatinine and lactate dehydrogenase did not return to normal range, which might be associated with the comorbidities of enrolled participants. However, some of the recovered patients still had the sequelae of shortness of breath, memory decay, and fatigue.
Table I.
Laboratory results, signs, and symptoms of convalescent patients with COVID-19
| Laboratory results, signs, and symptoms | Value (n = 78) |
|---|---|
| Average days from onset of illness | 381.00 (374.80-389.00) |
| Blood routine | |
| Leucocytes (×10⁹/L; normal range, 3.5-9.5) | 6.3 (1.6) |
| Increased | 2 (2.6%) |
| Decreased | 2 (2.6%) |
| Neutrophils (×10⁹/L; normal range, 1.8-6.3) | 3.6 (1.2) |
| Increased | 2 (2.6%) |
| Decreased | 1 (1.3%) |
| Lymphocytes (×10⁹/L; normal range, 1.1-3.2) | 2.1 (0.6) |
| Increased | 2 (2.6%) |
| Decreased | 1 (1.3%) |
| Platelets (×10⁹/L; normal range, 125.0-350.0) | 219.8 (52.5) |
| Increased | 1 (1.3%) |
| Decreased | 3 (3.8%) |
| Hemoglobin (g/L; normal range, male: 130.0-175.0; female: 115.0-150.0) | 140.8 (12.2) |
| Increased | 0 |
| Decreased | 0 |
| Liver function | |
| Total protein (g/L; normal range, 64.0-83.0) | 75.2 (4.4) |
| Decreased | 0 |
| Globulin (g/L; normal range, 20.0-35.0) | 30.4 (4.1) |
| Decreased | 0 |
| Albumin (g/L; normal range, 35.0-52.0) | 44.8 (1.8) |
| Decreased | 0 |
| Alanine aminotransferase (U/L; normal range, ≤41.0) | 22.7 (16.2) |
| Increased | 3 (3.8%) |
| Aspartate aminotransferase (U/L; normal range, ≤41.0) | 22.8 (8.6) |
| Increased | 2 (2.6%) |
| Renal function | |
| Blood urea nitrogen (mmol/L; normal range, 3.6-9.5) | 6.2 (1.4) |
| Increased | 1 (1.3%) |
| Decreased | 0 |
| Serum creatinine (μmol/L; normal range, male: 59-104; female: 45-84) | 77.3 (19.1) |
| Increased | 10 (12.8%) |
| Myocardial function | |
| cTnI (U/L; normal range, male: ≤34.2; female: ≤15.6) | 40.2 (23.4) |
| Increased | 2 (2.6%) |
| Lactate dehydrogenase (U/L; normal range, 135.0-225.0) | 215.2 (32.5) |
| Increased | 33 (42.3) |
| Infection-related biomarker | |
| IL-6 (pg/mL; normal range, 0.0-7.0) | 3.6 (2.5) |
| Increased | 4 (5.1%) |
| Signs and symptoms at detection | |
| Shortness of breath | 28 (35.9%) |
| Memory decay | 28 (35.9%) |
| Fatigue | 14 (17.9%) |
| Cough | 8 (10.3%) |
| Laryngeal discomfort | 8 (10.3%) |
| Diarrhea | 6 (7.7%) |
| Hypoacusis | 6 (1.7%) |
| Inattention | 6 (7.7%) |
| Expectoration | 4 (5.1%) |
| Muscle ache | 4 (5.1%) |
| Chest pain | 4 (5.1%) |
| Dizziness | 2 (2.6%) |
| Hypogeusia | 2 (2.6%) |
Data are presented as median (25th-75th), mean ± SD, or numbers (%). Increased means over the upper limit of the normal range and decreased means below the lower limit of the normal range.
Levels of anti-RBD, anti-N antibodies, and nAbs at 1 year postinfection
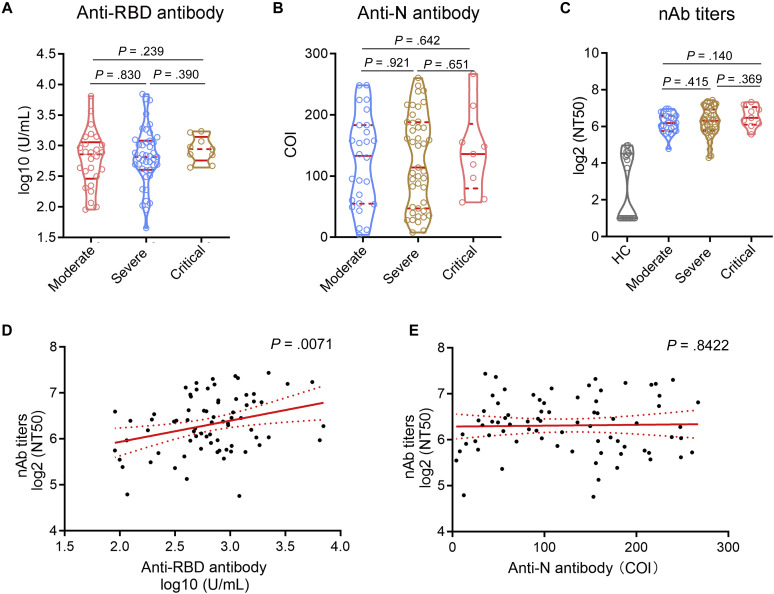
The positive rates of both anti-RBD and anti-N antibodies (total immunoglobulin) were 100% in convalescent patients after 1 year of recovery. Although previous studies have demonstrated a significantly higher IgG level in severe patients compared with moderate patients, either at the time of onset or at 6 months postinfection,15 , 27 we did not observe a statistical difference in both anti-RBD and anti-N antibody levels among different groups of patients at 1 year postinfection (Fig 1 , A and B).
Fig 1.
Levels of anti-RBD, anti-N antibodies, and nAbs. Levels of anti-RBD, anti-N antibodies, and nAb titers were detected in convalescent patients with COVID-19 (moderate, n = 26; severe, n = 43; critical, n = 9) at 1 year postinfection. A, Levels of anti-RBD antibodies in different groups were reported as concentrations (U/mL) and expressed as median with IQR (data were log10transformed). B, Levels of anti-N antibodies in different groups were reported as COI and expressed as median with IQR. C, nAb titers in HCs and different convalescent groups were reported as the log2 (NT50) and expressed as median with IQR. D, Correlation between nAb titers and anti-RBD antibody levels. E, Correlation between nAb titers and anti-N antibody levels. COI, Cutoff index; IQR, interquartile range; NT50, half-maximal neutralizing titer; HC, healthy control.
Consistent with anti-RBD and anti-N antibodies, the prevalence of nAbs also reached 93.59% in convalescent patients at 1 year postinfection. However, the nAb titers were significantly decreased in convalescent patients at 1 year postinfection compared with mild infections at the time of diagnosis (data of mild infections were reported in our previous study).14 Although there were progressive increases in nAb titers in convalescent patients with increasing severity of COVID-19, we did not observe a statistical difference among different groups (Fig 1, C). Notably, there was a significant correlation between nAb titers and anti-RBD antibody levels in convalescent patients with COVID-19 at 1 year postinfection, but anti-N antibody levels did not correlate with nAb titers, suggesting that anti-RBD antibody levels might be predictive of serum neutralization capabilities in patients with COVID-19 (Fig 1, D and E).
SARS-CoV-2–specific T-cell responses in convalescent patients at 1 year postinfection
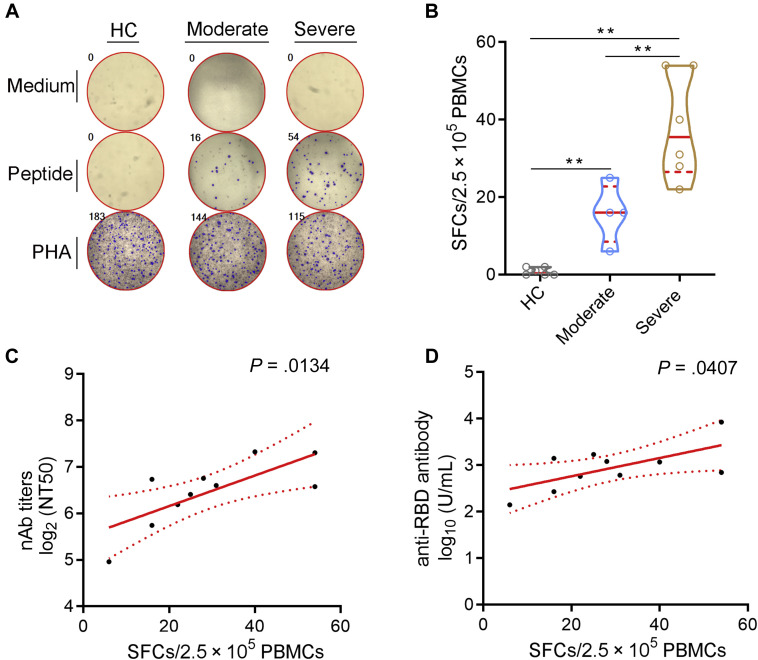
To gain a comprehensive insight into the immune memory to SARS-CoV-2, the frequency of viral-specific T cells was detected by using SARS-CoV-2–specific ELISPOT assay (Fig 2 , A). Surprisingly, after SARS-CoV-2 peptide pool stimulation, SARS-CoV-2–specific T cells were noted in 100% (10 of 10) of convalescent patients with COVID-19 after 1 year of recovery. Unlike nAbs, the number of SARS-CoV-2–specific T cells in severe convalescent patients was higher than that in moderate convalescent patients (Fig 2, B). Notably, the frequency of SARS-CoV-2–specific T cells was significantly positively correlated with nAb titers and anti-RBD antibody levels (Fig 2, C and D). These data confirmed the robust persistence of SARS-CoV-2–specific T-cell responses in convalescent patients with COVID-19 at 1 year postinfection.
Fig 2.
SARS-CoV-2–specific ELISPOT assay. PBMCs were isolated from convalescent patients at 1 year postinfection and stimulated with medium, SARS-CoV-2 peptide pool, or PHA for 18 to 24 hours. Then, the number of SARS-CoV-2–specific T cells was measured by IFN-γ ELISPOT assay. A, Representative graphs showing the numbers of SARS-CoV-2–specific T cells in HCs and convalescent patients with COVID-19 with moderate or severe disease. B, The numbers of SARS-CoV-2–specific T cells (SFCs/2.5 × 105 PBMCs) in different groups were expressed as median with IQR. C, Correlation between SARS-CoV-2–specific T-cell numbers and nAb titers. D, Correlation between SARS-CoV-2–specific T-cell numbers and anti-RBD antibody levels. HC, Healthy control; IQR, interquartile range; SFC, spot-forming cell. ∗∗P < .01.
The function and phenotype analysis of SARS-CoV-2–specific CD4+ T cells
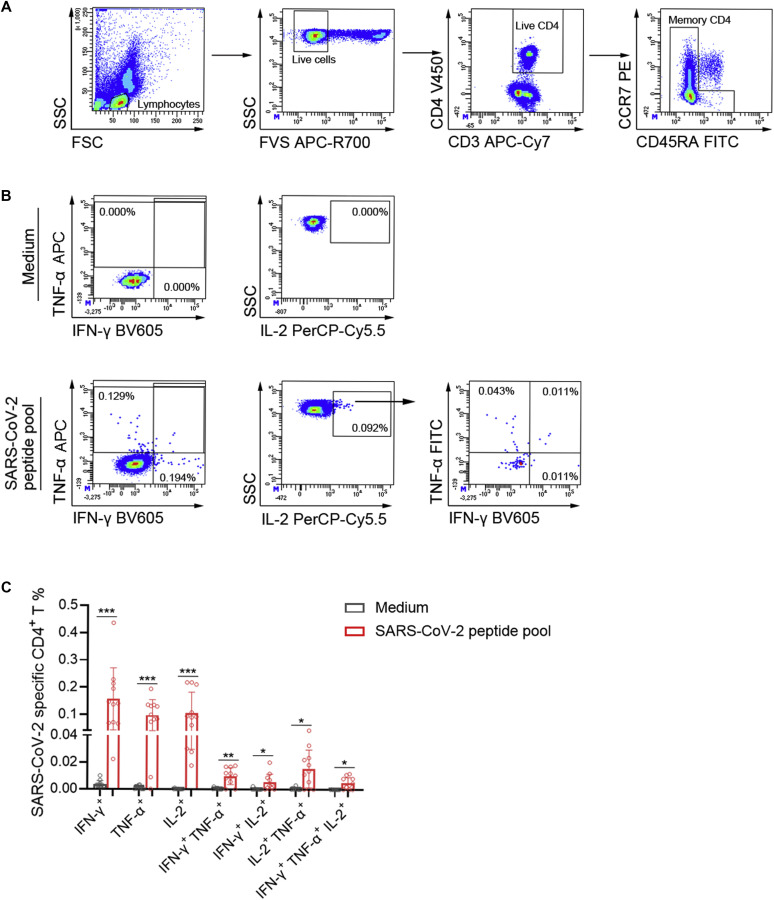
On peptide pool stimulation, IFN-γ+ SARS-CoV-2–specific CD4+ T cells were predominant effector memory cells (non-CD45RA+CCR7+ cells) (Fig 3 , A). After stimulation, amplified CD4+ T cells mainly produced a single cytokine (IFN-γ, TNF-α, or IL-2), whereas the percentage of multifunctional CD4+ T cells (IFN-γ+TNF-α+ or IFN-γ+TNF-α+IL-2+) was obviously lower than that of single cytokine-producing CD4+ T cells (Fig 3, B and C). Considering that SARS-CoV-2–specific CD4+ T cells mainly showed multifunctionality (expressing both IFN-γ and TNF-α) in patients with short-term recovery (∼3-8 weeks after the end of symptoms),28 these data suggested that SARS-CoV-2–specific CD4+ T cells were gradually losing their functional potential in convalescent patients with COVID-19 after 1 year of recovery.
Fig 3.
Cytokine secretion of SARS-CoV-2–specific CD4+ T cells. PBMCs isolated from convalescent patients at 1 year postinfection were stimulated with medium or SARS-CoV-2 peptide pool, and the percentages of IFN-γ+, TNF-α+, and IL-2+ cells within CD4+ T cells were analyzed. A and B, Representative flow dot plots showing the gating strategies of intracellular IFN-γ, TNF-α, and IL-2 in CD4+ T cells. Live lymphocytes were gated as FVS-negative cells. CD4+ T cells were selected as CD3+CD4+ T cells. Effector memory CD4+ T cells were gated by elimination of CCR7+CD45RA+ CD4+ T cells. B, IFN-γ+, TNF-α+, and IFN-γ+TNF-α+ cells in effector memory CD4+ T cells were gated. IL-2+ cells were gated from effector memory CD4+ T cells for analysis of IL-2+IFN-γ+, IL-2+TNF-α+, and IL-2+IFN-γ+TNF-α+ cells. C, The percentages of IFN-γ, TNF-α, and IL-2 single-, double- and triple-positive cells within CD4+ T cells in medium and SARS-CoV-2 peptide pool groups were expressed as mean ± SD. FITC, Fluorescein isothiocyanate; FSC, forward scatter; FVS, fixable viability stain; PE, phycoerythrin; SSC, side scatter.
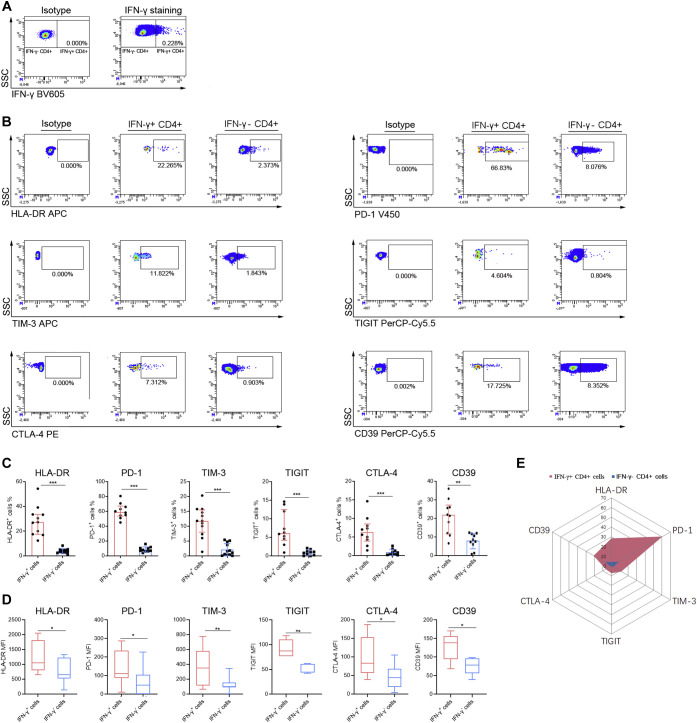
To further describe the characteristics of SARS-CoV-2–specific CD4+ T cells, cell phenotypes were analyzed by gating IFN-γ+CD4+ T cells after peptide pool stimulation (Fig 4 , A and B). We found that IFN-γ+ SARS-CoV-2–specific CD4+ T cells displayed both significantly higher percentages and mean fluorescence intensity of activation marker (HLA-DR) and exhaustion markers (PD-1, Tim-3, TIGIT, CTLA-4, and CD39) compared with IFN-γ−CD4+ T cells (Fig 4, C-E).
Fig 4.
Phenotype analysis of SARS-CoV-2–specific CD4+ T cells. PBMCs isolated from convalescent patients at 1 year postinfection were stimulated with SARS-CoV-2 peptide pool. After culture, the cells were collected for flow cytometry analysis. A, Representative flow dot plots showing the gating of IFN-γ+ and IFN-γ− CD4+ T cells. B, Representative flow dot plots showing the expressions of HLA-DR, PD-1, TIM-3, TIGIT, CTLA-4, and CD39 in IFN-γ+ and IFN-γ− CD4+ T cells. C, The percentages of HLA-DR, PD-1, TIM-3, TIGIT, CTLA-4, and CD39-positive cells in IFN-γ+ and IFN-γ− CD4+ T cells were expressed as mean ± SD. D, The MFIs of HLA-DR, PD-1, TIM-3, TIGIT, CTLA-4, and CD39-positive cells in IFN-γ+ and IFN-γ− CD4+ T cells were expressed as box plots (median, interquartile range, and min/max). E, Radar map analysis of the mean percentages of HLA-DR, PD-1, TIM-3, TIGIT, CTLA-4, and CD39-positive cells in IFN-γ+ and IFN-γ− CD4+ T cells. MFI, Mean fluorescence intensity; SSC, side scatter. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
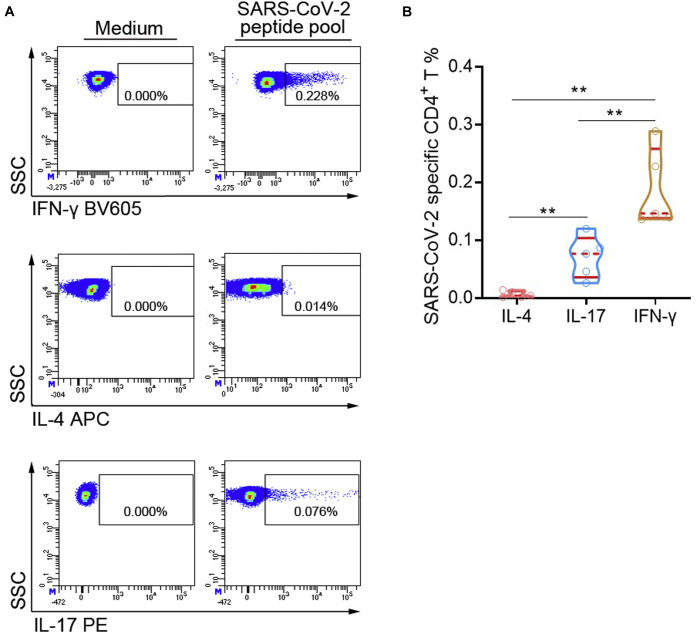
The subsets of SARS-CoV-2–specific CD4+ T cells were also determined according to the production of intracellular cytokines including IFN-γ (TH1), IL-4 (TH2), and IL-17 (TH17) (Fig 5 , A).29 After peptide pool stimulation, the percentages of IFN-γ+CD4+ TH1 cells were significantly higher than those of IL-17+CD4+ TH17 cells. However, we almost did not observe the differentiation of IL-4+CD4+ TH2 cells under peptide pool stimulation (Fig 5, B). Thus, SARS-CoV-2–specific CD4+ T cells were mainly differentiated into TH1 and TH17 cells, but not TH2 cells.
Fig 5.
The subsets of SARS-CoV-2–specific CD4+ T cells. PBMCs isolated from convalescent patients at 1 year postinfection were stimulated with SARS-CoV-2 peptide pool. After culture, the cells were collected for flow cytometry analysis. A, Representative flow dot plots showing the expressions of IFN-γ, IL-4, and IL-17 in CD4+ T cells. B, The percentages of IFN-γ+, IL-4+, and IL-17+ cells in CD4+ T cells were expressed as median with IQR. APC, Allophycocyanin; IQR, interquartile range; PE, phycoerythrin; SSC, side scatter. ∗∗P < .01.
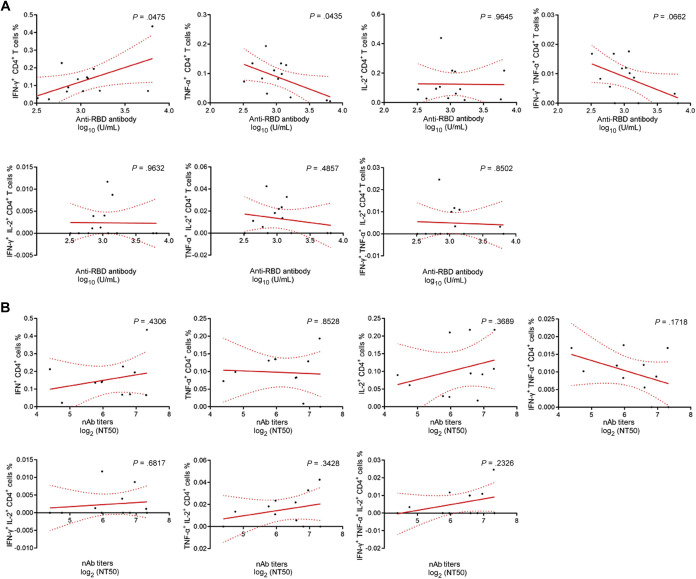
Moreover, after peptide pool stimulation, the percentage of IFN-γ– and TNF-α–producing CD4+ T cells was positively and negatively correlated with anti-RBD antibody levels, respectively. However, we did not observe a statistical correlation between the percentages of other cytokine-producing CD4+ T cells and antibody levels (Fig 6 ).
Fig 6.
Correlation analysis between different cytokine-producing CD4+ T cells and antibody levels. A, Correlation between the percentages of IFN-γ, TNF-α, and IL-2 single-, double-, and triple-positive cells within CD4+ T cells and anti-RBD antibody levels. B, Correlation between the percentages of IFN-γ, TNF-α, and IL-2 single-, double-, and triple-positive cells within CD4+ T cells and nAb titers.
Discussion
COVID-19 has become the greatest threat to global public health, and there is still no confirmed therapeutic strategy for the disease. How long the adaptive immunity triggered by SARS-CoV-2 can last is of critical clinical relevance in assessing the probability of second infection and efficacy of vaccination. Thus, the development of successful vaccination strategies depends on further understanding of the mechanism of immune memory. In this study, we confirmed that robust SARS-CoV-2–specific humoral and cellular responses were still maintained in convalescent patients with COVID-19 at 1 year postinfection, especially in those with more severe disease. Unfortunately, low nAb titers accompanied with exhausted function of SARS-CoV-2–specific CD4+ T cells indicated the gradual loss of immune memory to SARS-CoV-2 in convalescent patients at 1 year postinfection.
SARS-CoV-2–specific antibodies, especially the nAbs, can effectively curtail infection, based on blocking viral attachment and/or entry of host cells.30, 31, 32 A previous study has reported that both the proportion of participants with positive nAbs and the nAb titers are relatively stable for at least 9 months after SARS-CoV-2 infection, regardless of whether the individuals are symptomatic or not.27 Conversely, increasing evidences support the notion that nAb titers decrease significantly over 3 months and most convalescent patients with COVID-19 are reported to have low levels of nAbs.11 , 12 Discrepancy in the 2 results may be due to patients with different severity, because the magnitude of nAb titers is dependent on disease severity.12 Consistent with the later results, we observed that although the prevalence of nAbs in convalescent patients at 1 year postinfection was still high, the nAb titers in those patients after 1 year of recovery were relatively low compared with the titers in mild COVID-19 infections reported in our previous literature.14 Declining levels of nAbs may not be protective against COVID-19, as has been shown in the case of measles.33 Thus, although the definite protective activity of nAbs in patients with COVID-19 at 1 year postinfection remains unclear, the current data indicate that the protective role of nAbs is not optimistic because of low titers. But interestingly, anti-RBD antibody rather than anti-N antibody levels correlated well with both nAb titers and frequency of SARS-CoV-2–specific T cells, which indicated that commercially available anti-RBD antibody results can serve as useful surrogates for nAb testing and antigen-specific cellular responses. Furthermore, given that type I interferon immunity is essential for protective immunity to respiratory infection with SARS-CoV-2, a very recent study has indicated that autoantibodies neutralizing type I interferons predate SARS-CoV-2 infection and cause critical COVID-19, especially in the elderly.34 Thus, it is noteworthy that unlike nAbs, some types of antibodies in peripheral blood may not provide protection from infection, but conversely exacerbate illness.
In addition to nAbs, viral-specific T cells contribute to clearance of the acute infection.35 Among them, the most important ones are CD4+ TH cells, which orchestrate the immune responses and enable B cells to produce antibodies. A failure to develop protective immunity could occur because of T-cell response of insufficient magnitude or durability, because the nAb responses are dependent on the CD4+ T-cell responses.36 , 37 Accordingly, SARS-CoV-2–specific memory CD4+ T cells are noted in most convalescent patients with COVID and can persist for up to 6 to 7 months postinfection.22 , 26 Consistent with this notion, we observed that robust SARS-CoV-2–specific T-cell responses were maintained in convalescent patients with COVID-19 even at 1 year postinfection, in agreement with the status of humoral responses. These data confirmed that SARS-CoV-2–specific T and B cells could be retained long-term as populations of memory cells in convalescent patients with COVID-19.
The antigen-specific immunity depends on both the frequency and function of memory cells. We therefore further assessed the cytokine secretion ability and phenotypes of SARS-CoV-2–specific CD4+ T cells. To our surprise, on peptide pool stimulation, SARS-CoV-2–specific CD4+ T cells mainly produced a single cytokine, such as IFN-γ, TNF-α, and IL-2, which is inconsistent with a previous study that reported that amplified CD4+ T cells often showed multifunctionality (expressing both IFN-γ and TNF-α) in patients with short-term recovery.28 The different sampling intervals might cause such disparity. These data suggested that SARS-CoV-2–specific CD4+ T cells displayed strong function in short-term recovered patients but gradually lose their function in those individuals after 1 year of recovery. Consistent with this notion, we observed that the expressions of inhibitory receptors including PD-1, Tim-3, TIGIT, CTLA-4, and CD39 were all remarkably increased on SARS-CoV-2–specific CD4+ T cells. These findings indicated that although the number of SARS-CoV-2–specific CD4+ T cells was maintained in convalescent patients with COVID-19 after 1 year of recovery, their function may be exhausted.
Regarding PD-1 expression, a study found that PD-1–expressing SARS-CoV-2–specific CD8+ T cells were not exhausted, but functional in patients with COVID-19.38 This is in accordance with our previous study showing that increased expression of PD-1 on CD4+ T cells is correlated with higher cytokine secretion capability after phorbol 12-myristate 13-acetate/ionomycin stimulation.39 However, this study demonstrated that PD-1–expressing SARS-CoV-2–specific CD4+ T cells were exhausted but not multifunctional, and this discrepancy may be due to different detection periods. Previous studies assessed the role of PD-1 expression in patients with COVID-19 with active disease or in the short-term recovery period (within and after the first 14 days following the negative conversion of SARS-CoV-2 RNA), whereas this study focused on PD-1 expression in recovered patients at 1 year postinfection. Thus, PD-1 acts as an activation marker and expresses on effector SARS-CoV-2–specific T cells in the initial phase of disease, but over time it might gradually act as an exhaustion marker in the long-term recovery phase.
Several limitations of this study should be mentioned. First, although our data showed that the durability of humoral responses against SARS-CoV-2 in symptomatic patients was more than 1 year, the definite protective activity of nAbs needed to be further clarified. Second, we observed that robust SARS-CoV-2–specific T-cell responses were maintained in convalescent patients at 1 year postinfection, but we did not figure out which peptide these antigen-specific cells target. A further understanding of which SARS-CoV-2 peptide-specific T cells can maintain over a long-term period is crucial for vaccine design strategies. Third, this was a cross-sectional study performed in 1 center, and longitudinal data were not obtained in these patients.
Collectively, for the first time, our study provides evidence that robust SARS-CoV-2–specific humoral and cellular immunity is still maintained in convalescent patients with COVID-19 at 1 year postinfection. However, the loss of multifunctionality of SARS-CoV-2–specific CD4+ T cells accompanied with increased expression of inhibitory receptors on them supports the notion that SARS-CoV-2–specific CD4+ T-cell responses may not persist for longer periods. Our study not only extends our understanding of the durability of immune memory in convalescent patients with COVID-19 but also may have implications that vaccination is needed in convalescent patients for preventing reinfection.
Clinical implications.
The immune memory to SARS-CoV-2 in convalescent patients may persist for as long as 1 year, but the dysfunction of memory cells supports that vaccination is needed for preventing reinfection.
Footnotes
This study was supported by the National Mega Project on Major Infectious Disease Prevention (grant no. 2017ZX10103005-007) and COVID-19 Emergency Project from Wuhan Science and Technology Bureau (grant no. 2020020401010096).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Appendix
Table E1.
Anti-RBD and anti-N antibody quality controls and variation coefficients
| Controls | Statistical parameters | Anti-RBD |
Anti-N |
||
|---|---|---|---|---|---|
| Date | Value (U/mL) | Date | Value (COI) | ||
| Internal positive quality controls∗ | 12.10.2020 | 4.79 | 12.10.2020 | 3.07 | |
| 13.10.2020 | 4.85 | 13.10.2020 | 3.01 | ||
| 14.10.2020 | 4.78 | 14.10.2020 | 3.04 | ||
| 15.10.2020 | 4.74 | 15.10.2020 | 3.06 | ||
| Mean | 4.79 | 3.05 | |||
| SD | 0.05 | 0.03 | |||
| Intralot variation coefficient | 0.95% | 0.87% | |||
| External positive quality controls† | 12.10.2020 | 9.08 | 12.10.2020 | 2.72 | |
| 13.10.2020 | 9.16 | 13.10.2020 | 2.74 | ||
| 14.10.2020 | 8.87 | 14.10.2020 | 2.75 | ||
| 15.10.2020 | 8.98 | 15.10.2020 | 2.76 | ||
| Mean | 9.02 | 2.74 | |||
| SD | 0.13 | 0.02 | |||
| Intralot variation coefficient | 1.39% | 0.62% | |||
| External negative quality controls | 12.10.2020 | <0.4 | 12.10.2020 | 0.09 | |
| 13.10.2020 | <0.4 | 13.10.2020 | 0.09 | ||
| 14.10.2020 | <0.4 | 14.10.2020 | 0.10 | ||
| 15.10.2020 | <0.4 | 15.10.2020 | 0.10 | ||
| Mean | — | 0.09 | |||
| SD | — | 0.00331 | |||
| Intralot variation coefficient | — | 3.49% | |||
COI, Cutoff index; N, nucleoprotein.
In-house diluted leftover serum sample with high antibody levels, allowing for interlot and intralot comparison (determination of the coefficient of variation).
Manufacturer’s positive and negative quality controls (Roche SARS-Cov2 S PreciControl) that need to be respectively between 7.21 and 13.4 U/mL and below 0.4 U/mL, for the run to be carried out.
Table E2.
Baseline characteristics and clinical information of 78 convalescent patients with COVID-19
| Characteristic | Moderate (n = 26) | Severe (n = 43) | Critical (n = 9) | P value∗ |
|---|---|---|---|---|
| Age (y) (median, 25th-75th) | 64.5 (59.50-70.25) | 63.0 (56.00-68.00) | 67.0 (60.50-71.00) | .236 |
| Sex | ||||
| Female | 18 (69.23) | 24 (55.81) | 4 (44.44) | .311 |
| Male | 8 (30.77) | 19 (44.18) | 5 (55.56) | |
| Signs and symptoms at illness onset | ||||
| Fever | 16 (61.54) | 25 (58.14) | 7 (77.78) | .710 |
| Cough | 17 (65.38) | 30 (69.77) | 4 (44.44) | .624 |
| Chest distress | 5 (19.23) | 12 (27.90) | 2 (22.22) | .410 |
| Diarrhea | 7 (26.92) | 8 (18.60) | 1 (11.11) | .446 |
| Fatigue | 2 (7.69) | 6 (13.95) | 3 (33.33) | .569 |
| Shortness of breath | 1 (3.85) | 7 (16.28) | 2 (22.22) | .327 |
| Nausea and vomiting | 1 (3.85) | 1 (2.32) | 1 (11.11) | .674 |
| Headache | 0 | 2 (4.65) | 0 | .221 |
| Comorbidities | ||||
| Hypertension | 10 (38.46) | 14 (32.56) | 6 (66.67) | .518 |
| Diabetes | 2 (7.69) | 10 (23.26) | 2 (22.22) | .111 |
| Cardiovascular disease | 3 (11.54) | 3 (6.98) | 0 | .483 |
| Malignancy | 0 | 2 (4.65) | 1 (11.11) | .386 |
| Cerebrovascular disease | 0 | 0 | 2 (22.22) | .733 |
| Chronic kidney disease | 1 (0.38) | 0 | 0 | .180 |
| Tuberculosis | 1 (0.38) | 0 | 0 | .180 |
Data are presented as median (25th-75th) or numbers (%).
P values indicate the differences among moderate, severe, and critical patients. P < .05 was considered statistically significant.
References
- 1.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Lancet Emerging understandings of 2019-nCoV. Lancet. 2020;395:311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao A.T., Gao C., Zhang S. Profile of specific antibodies to SARS-CoV-2: the first report. J Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou H., Wang T., Zhang B., Luo Y., Mao L., Wang F., et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunology. 2020;9 doi: 10.1002/cti2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Pan Z., Yue S., Yu F., Zhang J., Yang Y., et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct Target Ther. 2020;5:180. doi: 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei Q., Li Y., Hou H.Y., Wang F., Ouyang Z.Q., Zhang Y., et al. Antibody dynamics to SARS-CoV-2 in asymptomatic COVID-19 infections. Allergy. 2021;76:551–561. doi: 10.1111/all.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C., Yu X., Gao C., Zhang L., Zhai H., Hu Y., et al. Characterization of antibody responses to SARS-CoV-2 in convalescent COVID-19 patients. J Med Virol. 2021;93:2227–2233. doi: 10.1002/jmv.26646. [DOI] [PubMed] [Google Scholar]
- 16.Zeng C., Evans J.P., Pearson R., Qu P., Zheng Y.M., Robinson R.T., et al. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight. 2020;5 doi: 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luchsinger L.L., Ransegnola B.P., Jin D.K., Muecksch F., Weisblum Y., Bao W., et al. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID-19 patients. J Clin Microbiol. 2020;58:e02005–e02020. doi: 10.1128/JCM.02005-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosendahl Huber S., van Beek J., de Jonge J., Luytjes W., van Baarle D. T cell responses to viral infections—opportunities for peptide vaccination. Front Immunol. 2014;5:171. doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seder R.A., Darrah P.A., Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 21.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 24.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J Clin Invest. 2021;131 doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroemer M., Spehner L., Vettoretti L., Bouard A., Eberst G., Pili Floury S., et al. COVID-19 patients display distinct SARS-CoV-2 specific T-cell responses according to disease severity. J Infect. 2021;82:282–327. doi: 10.1016/j.jinf.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Y., Liu F., Xu X., Ling Y., Huang W., Zhu Z., et al. Durability of neutralizing antibodies and T-cell response post SARS-CoV-2 infection. Front Med. 2020;14:746–751. doi: 10.1007/s11684-020-0822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Z., Ren L., Yang J., Guo L., Feng L., Ma C., et al. Seroprevalence and humoral immune durability of anti-SARS-CoV-2 antibodies in Wuhan, China: a longitudinal, population-level, cross-sectional study. Lancet. 2021;397:1075–1084. doi: 10.1016/S0140-6736(21)00238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 29.Mousset C.M., Hobo W., Woestenenk R., Preijers F., Dolstra H., van der Waart A.B. Comprehensive phenotyping of T cells using flow cytometry. Cytometry A. 2019;95:647–654. doi: 10.1002/cyto.a.23724. [DOI] [PubMed] [Google Scholar]
- 30.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murin C.D., Wilson I.A., Ward A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat Microbiol. 2019;4:734–747. doi: 10.1038/s41564-019-0392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R.T., Markowitz L.E., Albrecht P., Stewart J.A., Mofenson L.M., Preblud S.R., et al. Measles antibody: reevaluation of protective titers. J Infect Dis. 1990;162:1036–1042. doi: 10.1093/infdis/162.5.1036. [DOI] [PubMed] [Google Scholar]
- 34.Bastard P., Gervais A., Voyer T., Rosain J., Philippot Q., Manry J., et al. Autoantibodies neutralizing type I IFNs are present in ∼ 4% of uninfected individuals over 70 years old and account for ∼ 20% of COVID-19 deaths. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sallusto F., Lanzavecchia A., Araki K., Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., et al. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rha M.S., Jeong H.W., Ko J.H., Choi S.J., Seo I.H., Lee J.S., et al. PD-1-expressing SARS-CoV-2-specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54:44–52.e3. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., Hou H., Luo Y., Tang G., Wu S., Huang M., et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5 doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]