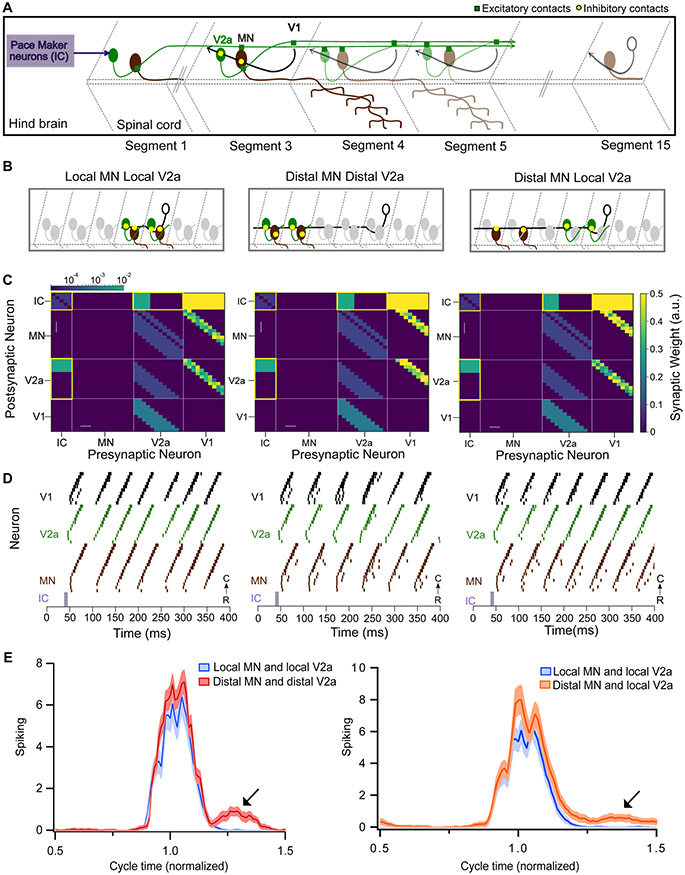
Figure 7: Modeling spinal circuitry with local and distal V1 inhibition.
A. Schematic of the computational model showing a reduced V1 (black), V2a (green), and motor neuron (MN, brown) network driven by rostrally located pacemaker neurons (purple box). B. Schematic of 3 different network structures simulated with the network model. Left: Local MN and local V2a connectivity from V1s. V1s synapse onto V2as and MNs located within 1 to 3 segments. Middle: Distal MN and distal V2a connectivity from V1s. V1s synapse onto V2as and MNs located 4 to 6 segments away. Right: Distal MNs and local V2a connectivity from V1s. V1s synapse onto MNs located 4 to 6 segments away and V2as located within 1 to 3 segments. C. Heatmap showing connectivity weights for neurons across 3 different network models. Connections highlighted in yellow are gap junctional and follow a logarithmic scale. Asterisks indicate altered connections. D. Raster plots of spike times from 1 representative simulation of each network. E. Spiking of MNs with respect to a normalized swim cycle. Black arrow points to the extraneous spiking observed for both distal MN / distal V2a connectivity (left) and more weakly in distal MNs / local V2a connectivity (right). See also Figure S7 and tables S1 and S2.