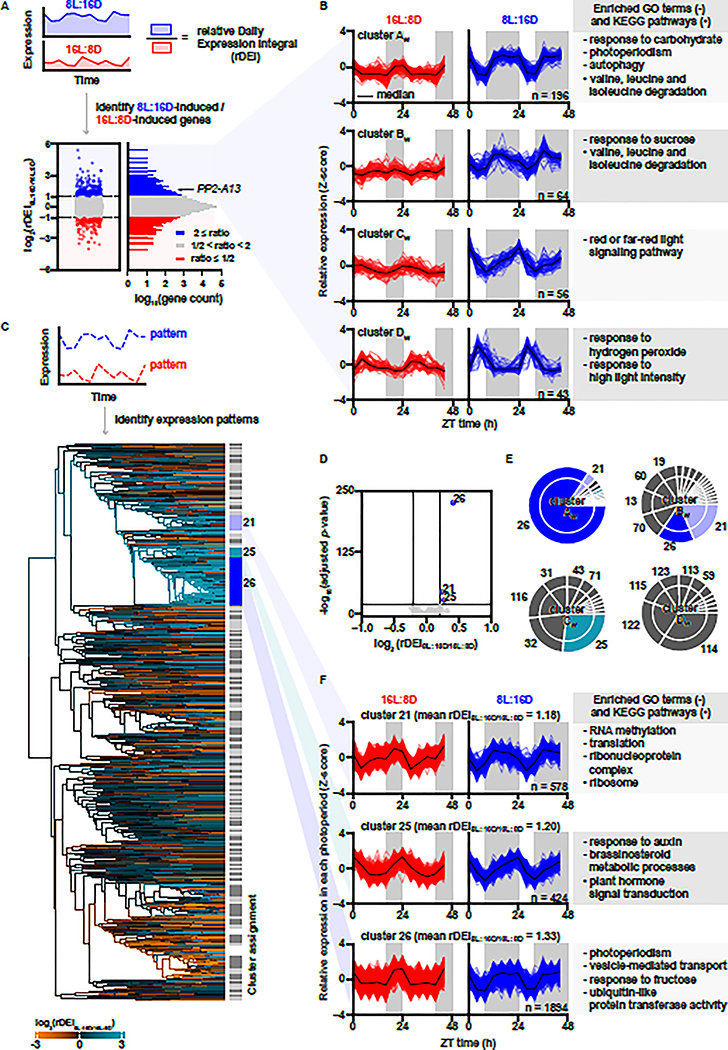
Figure 1. Induced gene expression in an 8L:16D photoperiod is correlated to rhythmic expression patterns with nighttime phasing.
(A) Distribution of rDEI8L:16D/16L:8D (n = 22810). Blue: 8L:16D-induced transcripts with rDEI8L:16D/16L:8D > 2.0; red: 16L:8D-induced transcripts with rDEI8L:16D/16L:8D < 0.5. (B) Normalized expression of 8L:16D-induced transcripts grouped by k-means clustering. Black lines indicate median expression level. Grey areas indicate darkness. Top enriched GO terms (−) and KEGG pathways (•) are listed. (C) Hierarchical clustering of all transcripts by 16L:8D and 8L:16D patterns (Table S4). Dendrogram edges are colored by the average rDEI8L:16D/16L:8D of transcripts in the node. Light grey and dark grey indicate clusters, and white indicates transcripts unassigned to any cluster. (D) Identification of photoperiod-induced clusters (average rDEI8L:16D/16L:8D > 1.15 or average rDEI8L:16D/16L:8D < 0.87; adjusted p-value < 1.0 × 10−20 one-sample Wilcoxon test with Bonferroni correction). Blue: 8L:16D-induced clusters. (E) Overlap between clusters of 8L:16D-induced transcripts and clusters of transcripts showing an 8L:16D-induction correlated pattern. (F) Expression pattern of transcripts in clusters 21, 25, and 26 normalized in each photoperiod (Table S4).