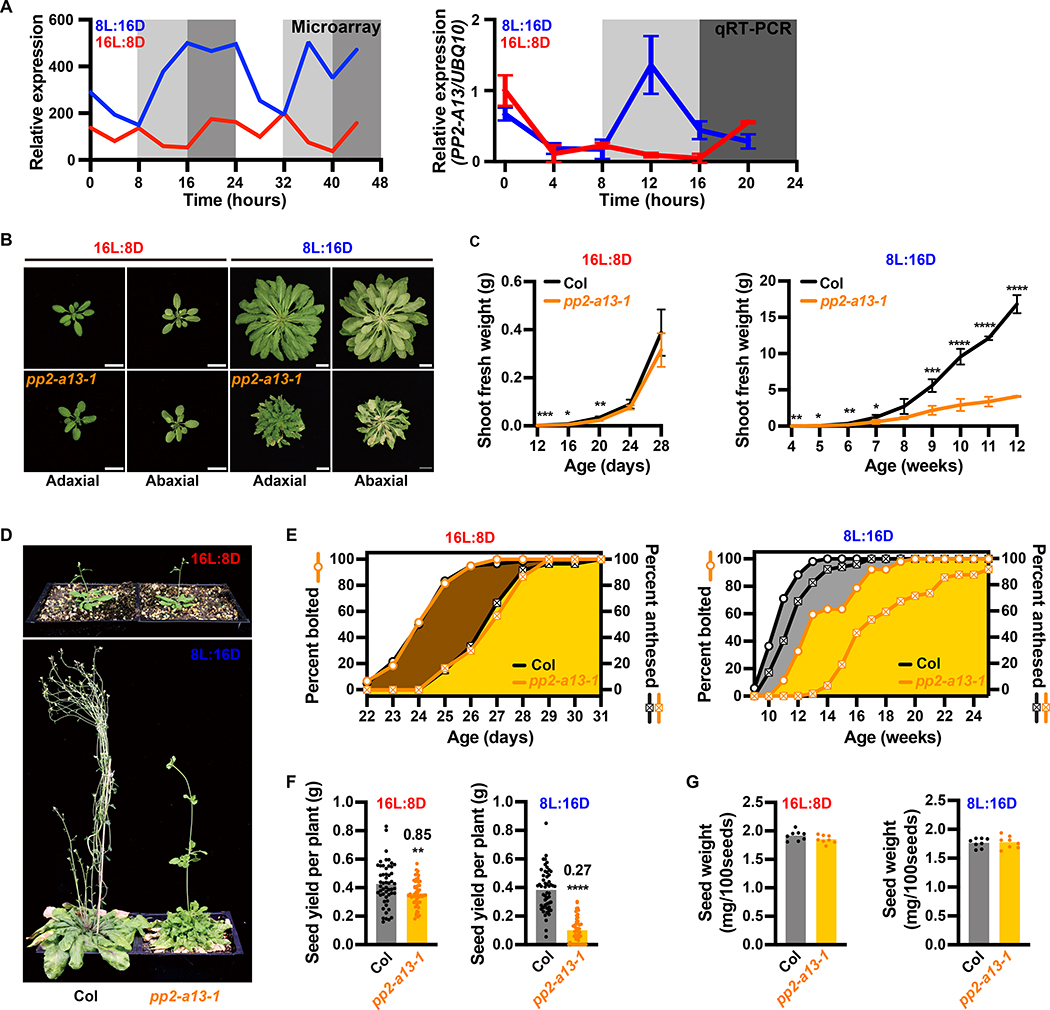
Figure 2. Disruption of the PP2-A13 gene causes 8L:16D-specific fitness defects.
(A) Microarray expression and qRT-PCR of PP2-A13 from 12-day-old plants grown in 8L:16D (blue) and 16L:8D (red). UBQ10 was used as an internal control. Error bars indicate SD, n = 3. (B) Representative wild type (Col) and pp2-a13–1 mutant plants grown for 24 days in 16L:8D or 11 weeks in 8L:16D. Adaxial and abaxial views of the rosettes are presented. Scale bar = 3 cm. (C) Aerial fresh weight of wild-type (Col, black) and pp2-a13–1 mutant (orange) plants grown in 16L:8D and 8L:16D. Error bar indicates SD. *, p⩽0.05; **, p⩽0.01; ***, p⩽0.001; ****, p⩽0.0001 (Welch’s t-test). (D) Representative wild-type (Col) and pp2-a13–1 mutant plants grown for 28 days in 16L:8D or 14 weeks in 8L:16D. (E) Percentage of wild-type (Col) and pp2-a13–1 mutant plants that are bolting or anthesed. Plants were grown in 16L:8D (left) and 8L:16D (right). n = 52–60. (F) Total seed yield from wild-type (Col) and pp2-a13–1 mutant plants grown in 8L:16D and 16L:8D. Numbers on the column indicate the fold change. n = 52–60. **, p⩽0.01; ****, p⩽0.0001 (Welch’s t-test). (G) Seed weight in mg/100 seeds from wild-type (Col) and pp2-a13–1 mutant plants grown in 8L:16D and 16L:8D. n = 8.