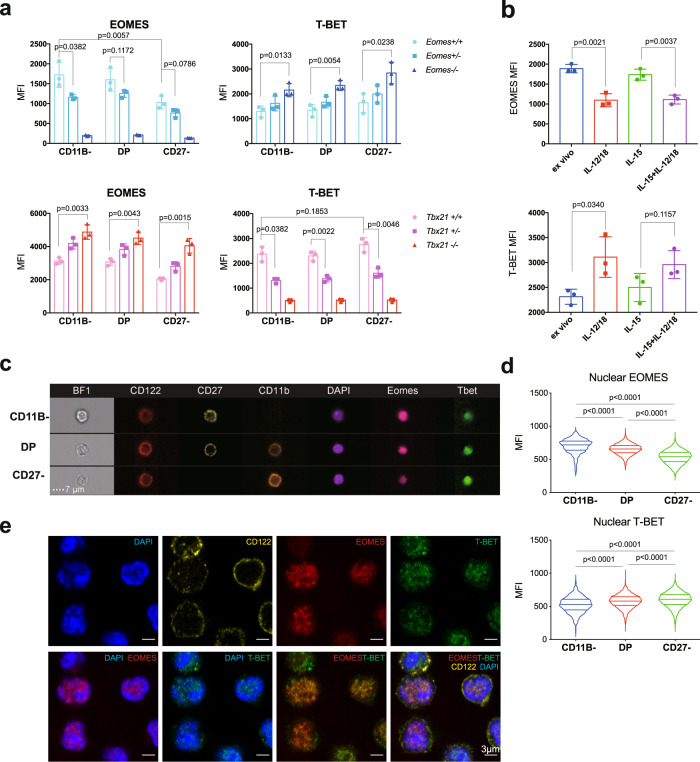
Fig. 1. EOMES and T-BET balance each other during NK cell maturation.
a Flow cytometry measurement of T-BET and EOMES in gated NK cells from WT, Tbx21+/−, Tbx21−/−, Ncr1Cre/+ (Eomes+/+), Ncr1Cre/+ X Eomeslox/+ (Eomes+/−) and Ncr1Cre/+ X Eomeslox/lox (Eomes−/−) mice, as indicated. Bar graphs show the mean ± SD fluorescence intensity (MFI) of EOMES and T-BET staining in gated NK cell subsets from spleen. Data are representative of 3 experiments and 3 mice are shown for each group. b WT spleen cells were stimulated O/N in the indicated conditions and T-BET and EOMES expression were measured by flow cytometry. Bar graphs show the mean ± SD fluorescence intensity (MFI) of EOMES and T-BET staining in gated NK cells. Data are representative of 2 experiments and 3 mice are shown for each group. c Image stream X (ISX) analysis of T-BET and EOMES expression in gated NK cells. Representative images are shown for the three NK cell subsets. CD122 staining was used to visualize the membrane. d Quantification of nuclear T-BET and EOMES in gated NK cell subsets analyzed with IDEAS software. Bar graphs show nuclear MFI of T-BET and EOMES for the indicated subsets, normalized to the level of each TF in immature CD11B- NK cells. N = 1232, 5157 and 22403 cells analyzed for CD11B-, DP and CD27- subsets, respectively. e NK cells were sorted from WT mice in immature or mature subsets and subsequently stained for nucleus (DAPI), T-BET, EOMES and CD122 for confocal microscopy analysis. Shown are representative images of each staining and combinations for immature NK cells. Images are representative from three individual experiments. Unpaired t tests (two-tailed) were used for statistical analysis of data presented in this figure.