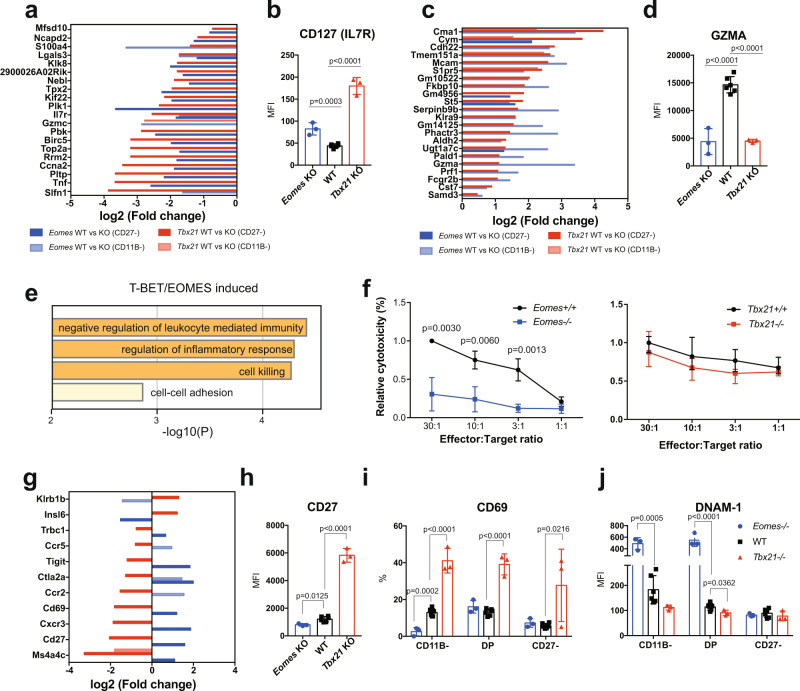
Fig. 5. Shared transcriptional activities of T-BET and EOMES in NK cells.
RNA-seq analysis of sorted Tbx21−/−, Eomes−/− and appropriate control CD11B- and CD27− NK cells (n = 3 per group with three sorts in total). DEGs were selected based on adjusted p < 0.05. a T-BET/EOMES co-repressed genes in immature or mature NK cells. Bar graphs show the log2 transformed fold change of genes between controls and T-BET or EOMES deficient mice, as indicated. b Bar graph showing the mean ± SD of IL7R expression in NK cells of the indicated genotype as measured by flow cytometry. The different controls for each genotype are pooled and annotated as “WT” in the graph which is same for the following graphs in (d, h–j). N = 3. c T-BET/EOMES co-induced genes in immature or mature NK cells, Bar graphs show the log2 transformed fold change of genes between controls and T-BET or EOMES deficient mice, as indicated. d Bar graph showing the mean ± SD of GZMA expression in NK cells of the indicated genotype as measured by flow cytometry. N = 3. e Functional annotation of the T-BET/EOMES induced gene set using Metascape. Bar graphs show selected terms among the most significant ones. f Cytotoxicity assay using sorted Tbx21−/−, Eomes−/− and appropriate control CD27− NK cells as effectors and RMA-KR-Nano-luc cells as targets. Graphs show the mean cytotoxicity ± SD. Data are from 3 mice in each group, pooled from two independent experiments. g Bar graph showing the log2 transformed fold change of genes regulated in opposite ways by T-BET and EOMES, between controls and T-BET or EOMES deficient mice, as indicated. N = 3–6. h–j Bar graphs showing the mean ± SD of CD27 (h), CD69 (i) and DNAM-1 (j) expression in NK cells of the indicated genotype as measured by flow cytometry. N = 3. Unpaired t tests (two-tailed) were used for statistical analysis of data presented in this figure.