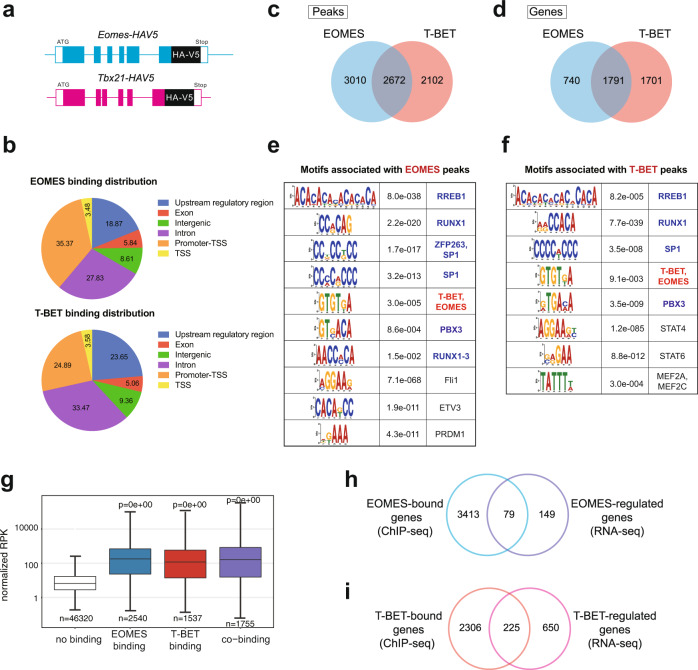
Fig. 8. Identification of direct T-BET and EOMES targets using novel KI mice.
a Scheme of knock-in Eomes and T-bet HAV5 constructs. b Distribution of EOMES and T-BET peaks relative to different genomic features. “Upstream regulatory regions” refers to regions located −20 kb/−1 Kb away from the TSS, “promoter-TSS” refers to regions located −1 Kb/+100 bp around the TSS and “TSS” refers to regions located −100 bp/+1 Kb around the TSS. c Venn diagram showing the overlap between EOMES and T-BET ChIP-seq peaks. d Venn diagram showing the overlap between genes nearby EOMES and T-BET ChIP-seq peaks. e–f Motifs associated with (e) EOMES or (f) T-BET peaks and corresponding TFs (E-values are shown, not ranked). g Boxplot presenting expression (RNA-seq data) of genes bound by T-BET and/or EOMES (Chip-seq data). Expression of genes without any of them is shown as reference. The number of genes analyzed is indicated in the figure. Minimal and maximal values are shown. The centre gives the mean expression and the box gives the 10–90 percentile. P values were calculated by Wilcoxon signed-rank tests. h Venn diagram showing the overlap between EOMES-bound (ChIP-seq) and EOMES-regulated genes (RNA-seq). i Venn diagram showing the overlap between T-BET-bound (ChIP-seq) and T-BET-regulated genes (RNA-seq). In panels (c, d, h, i) p values were calculated through hypergeometric tests and were p = 0; 0; 5.61133e-40; and 6.440556e-96, respectively.