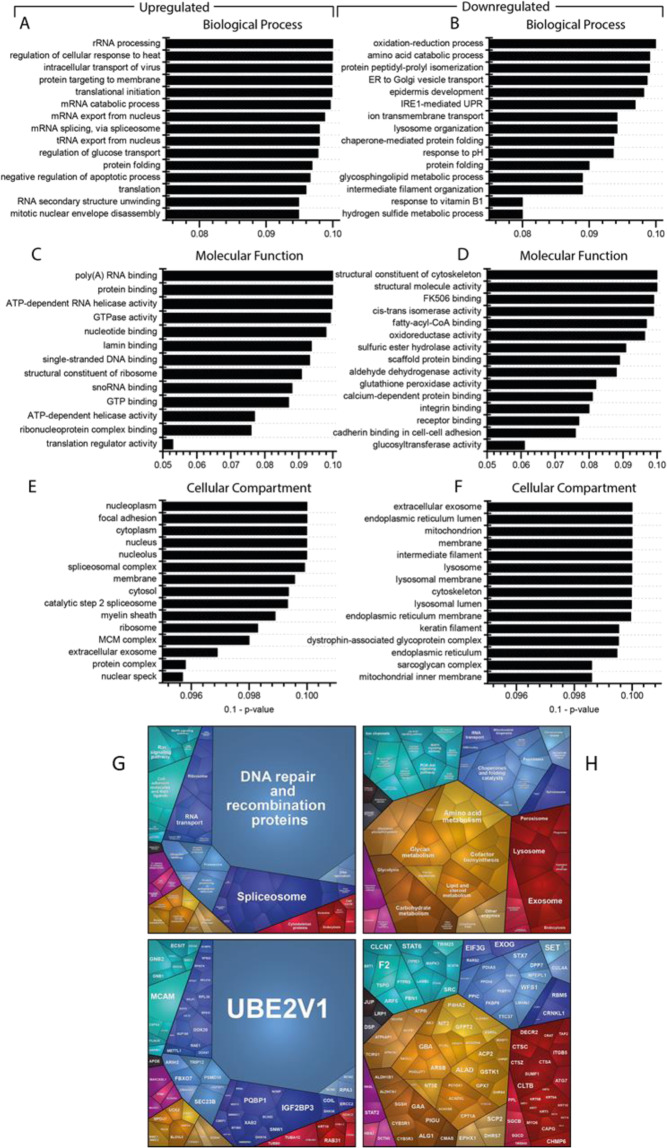
Fig. 2. In silico studies of proteomic data.
Gene Ontology (GO) term analysis was performed focussing on the up- and downregulated proteins separately: (A) “Biological Processes”revealed that the majority of increased proteins is involved in RNA processing, whereas most of decreased proteins (B) control the response to oxidative stress or are involved in protein folding. “Molecular Function” revealed that proteins involved in RNA-binding and modulating nucleotide binding are increased (C), and those involved in the cytoskeleton structure and the modulation of oxidative stress predominate amongst the downregulated (D). “Cellular Component” showed that the proteins upregulated in the patient-derived fibroblasts mostly belong to the nucleus (E) and the ER, the lysosome, mitochondria, and the cytoskeleton are downregulated (F). The proteomaps-based pathway analysis also focussed on increased and decreased proteins separately. G For increased proteins, it confirmed changes in RNA-transport and RNA-processing, and indicated an activation of the protein clearance machinery (Ubiquitin-mediated proteolysis via the proteasome) as well as altered signalling processes (Ras, ErbB and MAPK pathways). H For decreased proteins, indicated a vulnerability of cellular metabolism accompanied by perturbed signalling cascades (Jak-STAT, ErbB, MAPK, PI3K-AKT & NF-kappa-B pathways) and reduced lysosomal as well as exosome function.