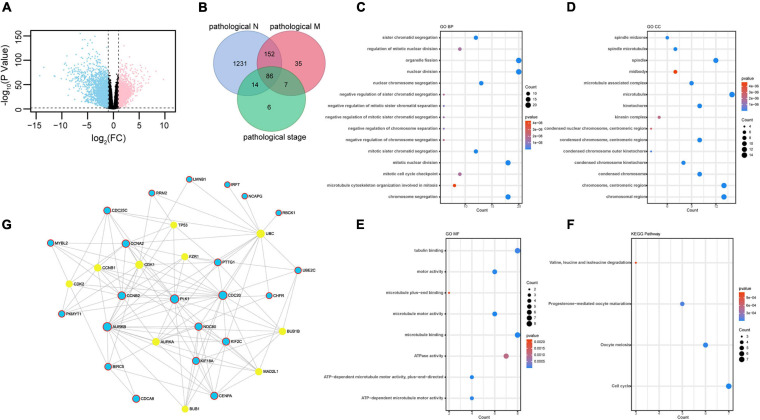
FIGURE 1.
Identification of differently expressed genes: (A) 4,933 DEGs identified in ccRCC TCGA dataset plotted in the volcano plot, in which the logarithmic ratio of FC of the tumor/normal are plotted against negative logarithmic P-values; 2,571 genes were significantly downregulated (blue); 2,362 genes were upregulated (red) (| fold change| > 2, p < 0.01). (B) Venn diagram of metastasis-related genes in ccRCC in TCGA dataset. A total of 86 genes showed significant association with pathological M (p < 0.05), pathological N (p < 1E-5), and pathological stage (p < 1E-10). (C–F) GO terms representing BP, CC, and MF and KEGG pathway analysis. (G) PPI network of 86 DEGs. The 22 seeds genes are shown in blue. The STRING Interactome database was selected to construct PPI, and the confidence score cutoff was set as 900; degree filter threshold values > 5.