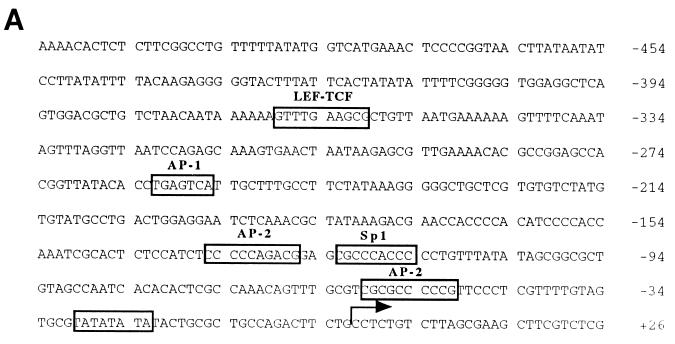
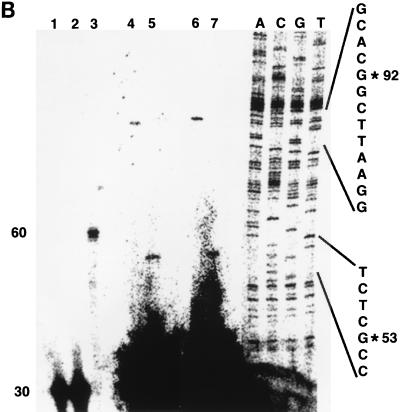
FIG. 3.
Determination of the transcription start site of the FN promoter. (A) Partial nucleotide sequence (−533/+27) of the genomic fibronectin clone XFN30.1. Numbering is with respect to the transcription start site (arrow). Conserved transcription factor binding sites are boxed. (B) Primer extension mapping of the 5′ termini of fibronectin transcripts. For primer extension, oligonucleotide FN-ext.3 (lane 1) or FN-ext.4 (lane 2), each 30 nucleotides in size, was labeled with T4 polynucleotide kinase. For size standards, either an unrelated oligonucleotide of 60 nucleotides was labeled (lane 3) or Sanger sequencing of an unrelated sequence of known composition was performed (lanes A, C, G, and T). Oligonucleotides were annealed to total CsCl-purified RNA from XTC cells and extended with MMLV reverse transcriptase. Two independent experiments are shown in lanes 4 and 5 and in lanes 6 and 7. Extension products of FN-ext.3 are shown in lanes 4 and 6, and those of FN-ext.4 are shown in lanes 5 and 7. RNA from lanes 6 and 7 was digested with DNase I prior to hybridization to exclude contamination with genomic DNA. No differences between the two experiments were detectable. In both cases, the extension product of FN-ext.3 was 92 nucleotides long, and that of FN-ext.4 was 53 nucleotides. None of the oligonucleotides annealed with tRNA (not shown).