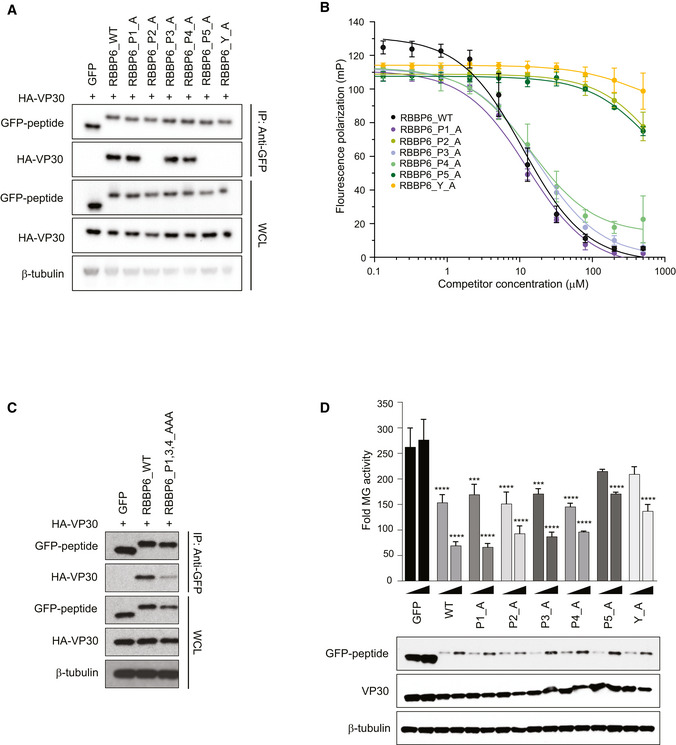
Co‐immunoprecipitation between HA‐VP30 and either GFP, or GFP‐fused to WT or mutant‐RBBP6 peptides. Each proline or tyrosine residue in the P1xP2P3P4P5xY motif was mutated to alanine, and the indicated mutant peptides were expressed as GFP fusions. Co‐immunoprecipitations of HA‐VP30 with GFP or GFP‐peptide fusions were performed using anti‐GFP magnetic beads (IP: Anti‐GFP). Whole cell lysates (WCL) were blotted with anti‐GFP, anti‐HA, and anti‐β‐tubulin antibodies.
Equilibrium dissociation curves of FITC‐RBBP6 from eVP30130‐272 as it is outcompeted by increasing concentrations (0.13–500 μM) of RBBP6 alanine mutants. Fluorescence polarization was determined with constant concentrations of FITC‐RBBP6 and eVP30130‐272 at 0.50 µM and 3.8 µM, respectively. Experiments were performed in two independent replicates. Error bars represent the standard deviation.
Co‐immunoprecipitation between HA‐VP30 and wild‐type (WT) or mutant‐RBBP6 peptides fused to GFP. GFP alone served as a negative control. IP was performed for GFP and blots were probed with anti‐GFP and anti‐HA antibodies. Anti‐β‐tubulin served as a loading control for the whole‐cell lysates.
Minigenome (MG) activity upon titration of plasmids encoding GFP fused to wild‐type or single point mutant‐RBBP6 peptides. GFP alone or GFP‐RBBP6 peptide plasmids were transfected at 12.5 and 25 ng amounts along with the plasmids of the MG system. The data represent the mean ± SD from one representative experiment in which each transfection condition was performed in triplicate (n = 3). The experiment was reproduced in two additional, independent experiments (see Appendix Fig S2). Statistical significance was calculated using ANOVA with Tukey’s multiple comparisons test. ****P < 0.00005; ***P < 0.0005