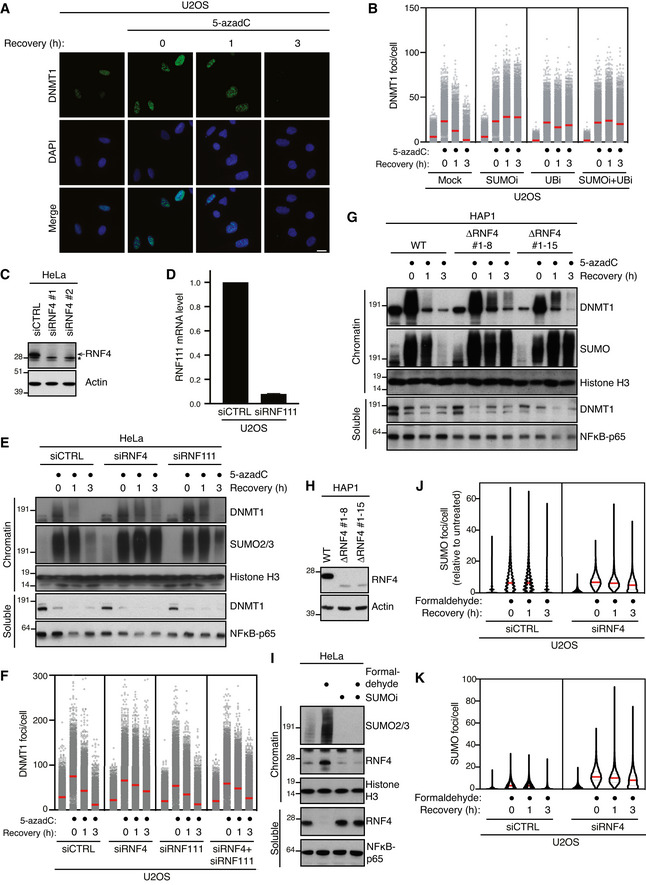
Representative images from the experiment shown in Fig
1B. Scale bar, 10 µm.
U2OS cells released from double thymidine synchronization in early S phase were pre‐treated or not with SUMOi for 15 min, pulse‐labelled with 5‐azadC for 30 min and incubated in the presence or absence of SUMOi and ubiquitin E1 inhibitor (UBi). Cells collected at the indicated times were pre‐extracted and immunostained with DNMT1 antibody. DNMT1 foci formation was analysed by quantitative image‐based cytometry (QIBC; red bars, mean; > 4,000 cells analysed per condition). Data are representative of three independent experiments.
Immunoblot analysis of HeLa cells transfected with non‐targeting control (CTRL) or RNF4 siRNA. *, cross‐reactive band.
U2OS cells were transfected with non‐targeting control (CTRL) or RNF111 siRNA. RNF111 mRNA levels were analysed by qPCR. Primers to GAPDH were used as a normalization control (mean ± SEM; n = 2 independent experiments).
HeLa cells transfected with indicated siRNAs were released from double thymidine synchronization in early S phase and treated with 5‐azadC for 30 min, washed and collected at the indicated times. Soluble and chromatin‐enriched fractions were immunoblotted with indicated antibodies.
U2OS cells transfected with indicated siRNAs were released from double thymidine block, pulse‐treated 2 h later with 5‐azadC for 30 min and processed for QIBC analysis of DNMT1 foci counts as in (B) (red bars, mean; > 8,500 cells analysed per condition). Data are representative of three independent experiments.
HAP1 cells (WT or independent derivative ΔRNF4 clones (#1–8 and #1–15) expressing a truncated form of RNF4 lacking the C‐terminal RING domain) were synchronized in S phase by overnight treatment with thymidine. Following removal of thymidine, cells were treated with 5‐azadC for 30 min and processed as in (E).
Immunoblot analysis of indicated HAP1 cell lines showing truncation and reduced expression of endogenous RNF4.
Immunoblot analysis of soluble and chromatin‐enriched fractions of HeLa cells treated with formaldehyde and/or SUMOi as indicated.
U2OS cells transfected with indicated siRNAs were treated with formaldehyde for 1 h. Cells were pre‐extracted and fixed at the indicated times after formaldehyde removal, immunostained with SUMO2/3 antibody and processed for QIBC analysis of SUMO2/3 foci counts (red bars, median; dashed lines, quartiles; > 6,000 cells analysed per condition). Data show SUMO2/3 foci counts per cell normalized to the mean foci count of untreated cells and are representative of three independent experiments.
As in (J), but showing total number of SUMO2/3 foci per cell.