Figure 2. DPC SUMOylation occurs in Xenopus egg extracts and is mediated by PIAS4.
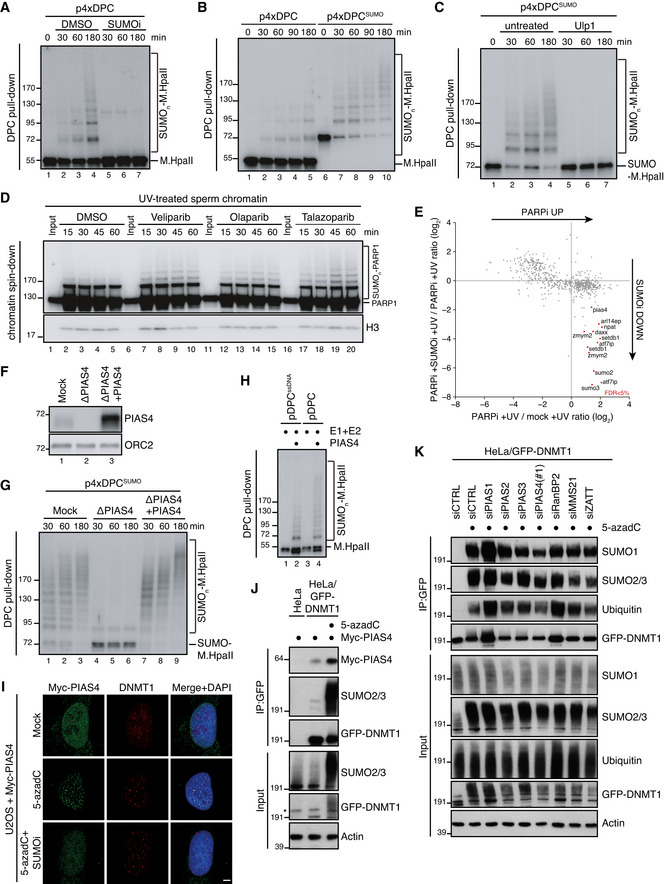
- M.HpaII was crosslinked into a plasmid to generate p4xDPC and then incubated in nucleoplasmic egg extracts (NPE) in the presence or absence of 50 µM of SUMOi. DPC pull‐down under stringent conditions was performed at the indicated time points, and the recovered samples were immunoblotted for crosslinked M.HpaII (Larsen et al, 2019). p4xDPC, which contains four M.HpaII DPCs, was used to increase sensitivity towards modified M.HpaII species. Identical results were obtained with plasmids containing one or four DPCs.
- p4xDPC or p4xDPCSUMO (generated by crosslinking SUMOΔGG‐M.HpaII) were incubated in NPE and DPC pull‐down performed as in (A). Note that priming M.HpaII with SUMO stimulates rapid poly‐SUMOylation of the DPC in NPE. Thus, this substrate was used in many subsequent experiments to stimulate DPC poly‐SUMOylation.
- p4xDPCSUMO was incubated in NPE and plasmids analysed as in (A). Following DPC pull‐down, the samples were split and either left untreated or treated with the SUMO protease Ulp1.
- Sperm chromatin treated with 2,000 J/m2 of UV‐C was incubated in non‐replicating egg extracts in the presence or absence of the indicated PARP inhibitors (PARPi; Veliparib (50 µM), Olaparib (50 µM), Talazoparib (10 µM)). At the indicated time points, chromatin was recovered via chromatin spin‐down and samples were immunoblotted with indicated antibodies.
- Plot illustrating protein recruitment to UV‐treated sperm chromatin in the presence or absence of Talazoparib and SUMOi, as determined by CHROMASS analysis (Dataset EV1). Red dots indicate the proteins that are significantly enriched on sperm chromatin in the presence of PARPi in a SUMO‐dependent manner (n = 4 biochemical replicates; FDR < 5% corresponds to a permutation‐based FDR‐adjusted q‐value of < 0.05). Note that different isoforms of the same protein (e.g. ATF7IP) can sometimes be detected.
- NPE was either mock‐ or PIAS4‐depleted, and recombinant xPIAS4 was added where indicated to a final concentration of 10 ng/µl. Protein samples were immunoblotted with the indicated antibodies.
- Samples from (F) were added to p4xDPCSUMO for the indicated times and recovered via DPC pull‐down as in (A).
- pDPC or a plasmid containing M.HpaII crosslinked to ssDNA (pDPCssDNA) (Larsen et al, 2019) was incubated with SUMO E1 and E2 enzymes and SUMO, in the presence or absence of recombinant xPIAS4. Samples were recovered by DPC pull‐down and blotted against M.HpaII as in (A).
- Representative images of U2OS cells transfected with Myc‐PIAS4 expression plasmid that were left untreated or exposed to 5‐azadC in the presence or absence of SUMOi, fixed 2 h later, pre‐extracted and co‐immunostained with DNMT1 and Myc antibodies. Scale bar, 5 µm.
- HeLa or HeLa/GFP‐DNMT1 cells left untreated or exposed to 5‐azadC for 30 min were lysed and subjected to GFP IP under stringent conditions. After extensive washing, individual IPs were incubated with an equal amount (800 µg) of whole cell lysate of HeLa cells transfected with Myc‐PIAS4 expression construct (Fig EV2J), washed and immunoblotted with antibodies to Myc, SUMO2/3 and GFP. *, cross‐reactive band.
- HeLa/GFP‐DNMT1 cells transfected with previously validated siRNAs targeting established SUMO E3 ligases (Fig EV2L and Methods section) were treated with 5‐azadC for 30 min, collected and subjected to GFP immunoprecipitation under denaturing conditions, and immunoblotted with antibodies to SUMO1, SUMO2/3, ubiquitin and GFP.
