Graphical abstract

Keywords: Antibiotic resistance, Polymyxin, LPS, Outer membrane, Molecular dynamics simulations
Highlights
-
•
A structural model of bacterial outer membrane (OM) was developed with Ra LPS.
-
•
Free energy landscape was revealed for polymyxin interactions with the OM.
-
•
LPS core sugars and calcium ions confer intrinsic resistance to antibiotics.
Abstract
The outer membrane (OM) of Gram-negative bacteria is a formidable barrier against antibiotics. Understanding the structure and function of the OM is essential for the discovery of novel membrane-acting agents against multidrug-resistant Gram-negative pathogens. However, it remains challenging to obtain three-dimensional structure of bacterial membranes using crystallographic approaches, which has significantly hindered the elucidation of its interaction with antibiotics. Here, we developed an asymmetric OM model consisting of rough lipopolysaccharide (LPS) and three key types of phospholipids. Using coarse-grained molecular dynamics simulations, we investigated the interaction dynamics of LPS-containing OM with the polymyxins, a last-line class of antibiotics against Gram-negative ‘superbugs’. We discovered that polymyxin molecules spontaneously penetrated the OM core sugar region where most were trapped before entering the lipid A region. Examination of the free energy profile of polymyxin penetration revealed a major free energy barrier at the LPS inner core and lipid A interface. Further analysis revealed calcium ions predominantly distributed in the inner core region and mediated extensive cross-linking interactions between LPS molecules, thereby inhibiting the penetration of polymyxins into the hydrophobic region of the OM. Collectively, our results provide novel mechanistic insights into an intrinsic defense of Gram-negative bacteria to polymyxins and may help identify new antimicrobial targets.
1. Introduction
Antibiotics are one of the greatest medical discoveries in the 20th century. However, over the last few decades bacterial isolates have developed resistance to most, and in some cases all currently available antibiotics. [1] Of particular concern are multidrug-resistant (MDR) Gram-negative pathogens such as Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae, which have caused significant morbidity and mortality. [2], [3] This situation has been made worse by the dry antibiotic development pipeline since the 1990 s, dramatically reducing the antibiotic armamentarium available to treat infections caused by these difficult-to-treat organisms. Therefore, the polymyxins (i.e., polymyxin B and colistin), an ‘old’ class of antibiotics once abandoned in the 1970 s due to toxicity, are increasingly used as a last-line therapy against these life-threatening ‘superbugs’. [4], [5]
Polymyxins are polycationic lipopeptides that act primarily through binding to lipopolysaccharide (LPS) and disrupting the outer membrane (OM) of Gram-negative bacteria. [6], [7] LPS is the major component in the outer leaflet of the bacterial OM and consists of a conserved lipid A moiety, a core oligosaccharide and an O-antigen. The direct interactions between adjacent LPS molecules and indirect interactions mediated by divalent cations significantly strengthen the OM structure, protecting bacterial cells from environmental toxins, including antibiotics. [8], [9] OM porins are responsible for the transport and uptake of many lower molecular weight polar antibiotics; [10] however, it is very likely that the small pore size (~1 nm) of such porins prohibits the translocation of larger (~2 nm) lipopeptides such as the polymyxins. Our recent all-atom molecular dynamics (MD) simulations have revealed that the antimicrobial activity of polymyxins was highly dependent on their specific interactions with lipid A and unique conformational folding within the bacterial OM. [6] Unfortunately, such activity-related prerequisites are mitigated when the lipid A phosphate groups are modified with phosphoethanolamine and/or 4-amino-4-deoxy-L-arabinose moieties. [11] These lipid A modifications are the main mechanisms of polymyxin resistance in Gram-negative bacteria. Even in the absence of such modifications, the core oligosaccharide of LPS restricts the insertion of polymyxins into the OM. [12], [13] Removal of these core sugars from LPS is known to improve the OM penetration of polymyxins. [14] These findings indicate the role of LPS core sugars in contributing to polymyxin resistance. However, exploiting the interaction between polymyxins and the bacterial OM at the molecular level remains challenging with current experimental approaches. While a small number of MD simulation studies examined the action of polymyxins against the bacterial OM, [6], [11], [15] the core oligosaccharide section of LPS is often not included when constructing the OM models. This has hindered our understanding of the role of LPS core sugars in antibiotic resistance.
In the present study, we developed a bacterial OM model incorporating rough LPS (Ra LPS, consisting of lipid A and a complete oligosaccharide chain) and examined the interactions with polymyxin B1 at three different concentrations. To enhance the conformational sampling during the process of polymyxin interaction with the OM, we integrated coarse-grained (CG) simulations and free energy analysis to examine the thermodynamic mechanism governing the penetration of polymyxins into the OM. Together with the energy-unfavorable calcium displacement by polymyxins, the anionic LPS core sugars acted as an ‘energy trap’ to prevent the penetration of polymyxins into the lipid A region of the OM, which provided an intrinsic defense mechanism against polycationic polymyxins.
2. Methods
2.1. System preparation
We have previously developed and validated the three-dimensional atomic structure of polymyxin B1. [6], [11], [16] To build the CG model of polymyxin B1, approximately four heavy atoms were mapped to a single CG bead (Fig. 1A), consistent with the setting of Martini CG particle building blocks. [17] Subsequently, all-atom and CG MD simulations were performed for a single polymyxin B1 molecule in a 5 × 5 × 5 nm3 box (with neutralized sodium chloride) to calculate the polymyxin B1 molecular parameters (e.g., bonds and angles). [18] The CG model and parameters of Ra LPS (Fig. 1B) were from CHARMM-GUI and validated elsewhere. [19]
Fig. 1.
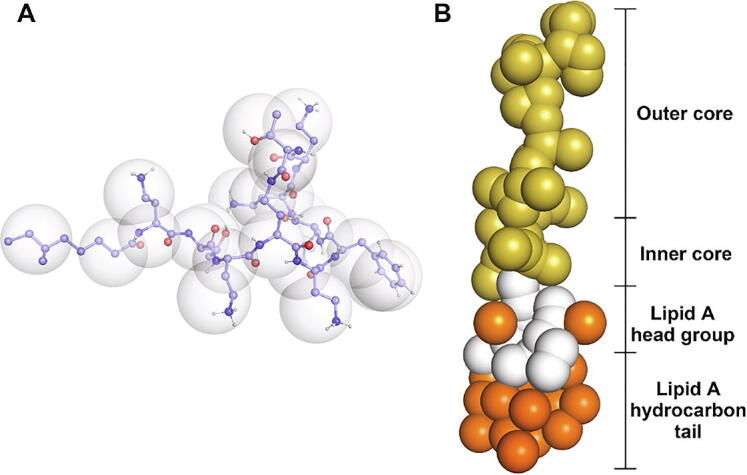
CG models for polymyxin B1 and Ra LPS. (A) CG model for polymyxin B1. The Martini beads are shown as transparent spheres, while the underlying united-atom particles are shown as spheres. Carbon atoms are light blue, oxygen atoms are red, nitrogen atoms are dark blue, and hydrogen atoms are white. (B) CG model for Ra LPS. Hydrocarbon tails are orange, the lipid A head group is white, phosphate groups are orange, and the remaining outer and inner core saccharides are yellow. The inner core indicates 2-keto-3-deoxtoctonoic acid (KDO). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
An asymmetric bacterial OM model was constructed using the CHARMM-GUI Martini Maker module. [19] The outer leaflet of the OM consisted of 96 Ra LPS molecules whereas the inner leaflet contained 80% 1-palmitoyl-2-oleoyl-phosphatidylethanolamine (POPE), 10% 1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG), and 10% cardiolipin (96 Ra LPS and 263 phospholipids in total). This represents the composition of the heterogeneous OM of Gram-negative bacteria. [6], [20] The OM model was then solvated using CG water particles and neutralized using calcium and chloride ions in a 13.1 × 13.1 × 16.3 nm3 simulation box. To investigate the effect of polymyxin concentration on interactions with the OM, 24, 48 and 96 polymyxin B1 molecules were added above the OM to achieve a 1:4, 1:2 and 1:1 polymyxin B1/LPS ratio (P/L ratio), respectively. As a result, the number of calcium ions in these systems was 420, 360 and 240, respectively. A control system was also performed in the absence of polymyxin molecules. In total, there were approximately 46,000 particles in the simulation systems.
2.2. MD simulations
All simulations were conducted with the GROMACS program (version 5.1.2) and Martini force field. [17], [21] The energy minimization was performed using the steepest decent algorithm. Subsequently, a six-step equilibration cycle was carried out by gradually turning off the position restraints on the lipid molecules (both LPS and phospholipid). Finally, 3,000-ns production simulations were conducted for each system in the NPT ensemble. In particular, 2,000-ns extended simulations were performed for the system with 96 polymyxin B1 molecules (1:1 P/L ratio). The temperature and pressure were coupled to 313 K using the velocity rescaling method (time constant of 1 ps) [22] and 1 bar using the semi-isotropic barostat and Parrinello-Rahman algorithm (time constant of 5 ps), [23] respectively. Other simulation details were consistent with previous validated studies. [18]
2.3. Umbrella sampling and free energy calculation
Compared to the systems with 1:2 and 1:4 P/L ratios, the polymyxin B1 molecules inserted most deeply into the OM in the 1:1 P/L ratio system (details introduced in the following sections). Therefore, umbrella sampling simulations [18], [24] were performed using the trajectories of the 1:1 P/L ratio system to elucidate the energetic mechanisms governing the dynamic behaviors of polymyxin molecules upon interacting with the OM. Snapshots from the 1:1 P/L ratio system were generated and employed to initiate 10 simulation windows in which the polymyxin B1 molecule was restrained at fixed positions with a 0.2-nm interval and 1,000 kJ mol−1 nm−2 force constant of harmonic potential. The spring force was applied only on the polymyxin molecule that showed the deepest insertion in the MD simulations and no restraints were applied on the other polymyxin molecules. Each window was simulated for 300 ns to enhance conformational sampling. Finally, the free energy profile was determined using the Weighted Histogram Analysis Method (WHAM) and displayed as a function of the absolute Z-coordinates of the system. [25] Other running parameters were set to match the unbiased MD simulations.
3. Results
3.1. Interaction of polymyxin B1 at various concentrations with the Ra LPS OM
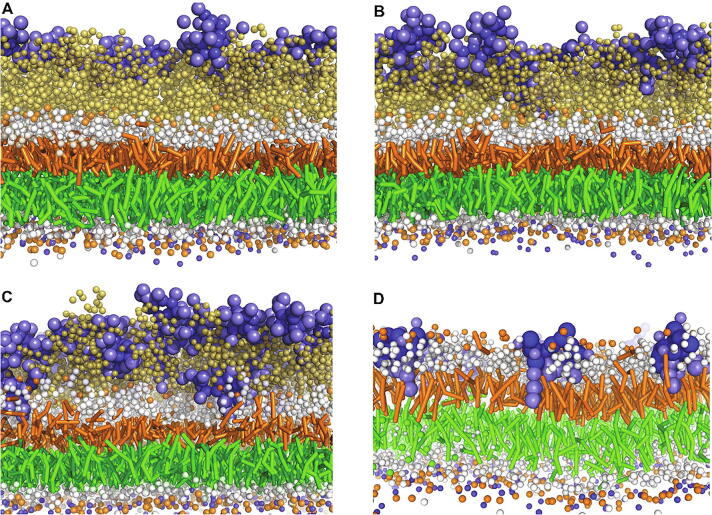
In the 1:4 P/L ratio system, approximately 60% (14 of 24) of polymyxin B1 molecules were trapped in the outer core of LPS, while the remaining molecules traversed the periodic boundary to interact with the OM inner leaflet or floated in water (Fig. 2A); no obvious penetration into the OM was observed throughout the entire 3,000-ns simulations. In the 1:2 P/L ratio system, only two polymyxin B1 molecules approached the inner core of LPS while the majority of molecules bound to the outer core region (Fig. 2B). As the polymyxin concentration further increased to a 1:1 P/L ratio, seven polymyxin B1 molecules passed the outer and inner core regions of LPS and interacted with the lipid A headgroups (Fig. 2C). However, these polymyxin B1 molecules did not enter the hydrophobic region of the OM. Interestingly, in simulations involving a new lipid A OM where the core sugars were removed from the Ra LPS, polymyxin B1 molecules easily inserted into the hydrophobic region of the lipid A OM, even at a very low concentration (1:18 P/L ratio) (Fig. 2D). These results show that the core sugars of LPS ‘trapped’ polymyxin molecules and inhibited their penetration into bacterial OM, most likely via electrostatic interactions between the negatively charged carboxyl groups of the inner core sugars and the positively charged 2,4-diaminobutyric acid (Dab) residues of polymyxin B1.
Fig. 2.
Final frame snapshots showing polymyxin B1 (blue) interacting with the Ra LPS outer membrane. (A) 1:4, (B) 1:2, (C) 1:1 polymyxin/LPS ratios, and (D) polymyxin B1 interaction with lipid A outer membrane with 1:18 polymyxin/lipid A ratio. Ra LPS and lipid A are colored as in Fig. 1B; phospholipids in the inner leaflet of the outer membrane are shown with green hydrocarbon tails and white head groups. The polymyxin B1 molecules across the simulation box are not shown. Na+, Ca2+ and water molecules are omitted for clarity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further investigate the dynamic behaviors of polymyxins within the OM, we examined the insertion kinetics of a single polymyxin molecule showing the deepest penetration in the 1:1 P/L ratio system (Fig. 3). This polymyxin B1 molecule moved rapidly from the surface of the OM to the interior of the outer core region (0–200 ns), before slowly approaching the inner core region (200–1,000 ns). Subsequently, it spent approximately 1,400 ns translocating through the inner core region (1,000–2,400 ns) before contacting the lipid A headgroups. For the remainder of the simulation (2,400–3,000 ns), it fluctuated within the inner core region of the OM. This remained the case even when the simulations were extended by 2,000 ns, with the molecule never inserting into the lipid A region. These results show that the core sugars of LPS likely constitute a permeability barrier to polycationic polymyxins.
Fig. 3.

Interaction of polymyxin B1 with the Ra LPS outer membrane in a 1:1 peptide/LPS system. Regions of Ra LPS leaflet of the outer membrane are indicated by different transparent colors. Positions of the center of mass of a representative polymyxin B1 molecule are shown as black lines. The coordinates are shown with respect to the bilayer normal.
3.2. Calcium ions within the OM
Given that divalent cations help stabilize the bacterial OM, we hypothesized that calcium ions may play a key role in maintaining the permeability barrier created by the LPS core sugars. To test this hypothesis, the calcium distribution within the OM was measured in the absence of polymyxin B1 and with three different P/L ratios (Fig. 4A). In each system, a major peak in the inner core region (around Z = 7.7 nm) and a minor peak in the outer core region (around Z = 8.7 nm) was observed. These calcium ions interacted electrostatically with the anionic sugars of LPS in the outer and inner cores (Fig. S1), thereby stabilizing the hydrophilic network between LPS. However, compared to the control system, the mass density of calcium ions in the inner core decreased by 20.2% and 29.1% with the 1:4 and 1:2 P/L systems, respectively, despite comparable levels in the outer core in both systems. In contrast, in the 1:1 P/L system the mass density dropped by 63.6% and 45.3% in the inner and outer core regions, respectively. These results provide new quantitative insights into the calcium displacement by polymyxins, which is consistent with a previous study. [26]
Fig. 4.
Distributions of calcium ions (A) within the Ra LPS outer membrane and (B) around the translocated polymyxin B1 molecule. The transparent colored shallows show the different regions of the Ra LPS leaflet of the outer membrane.
To examine the potential interaction between polymyxins and calcium ions, we measured the number of calcium ions around (within 1 nm) the inserted polymyxin molecule (Fig. 4B). During the insertion process, there were 4–5 calcium ions around the polymyxin molecule as it traversed the outer core region. However, less than one calcium ion was found around the polymyxin molecule when it subsequently entered the inner core region. Nevertheless, the inner core calcium ions showed the highest mass density (Fig. 4A). Given both polymyxins and calcium ions are positively charged, these results clearly suggest that penetration of polymyxins causes the expulsion of calcium ions from the OM, most likely due to electrostatic repulsion.
3.3. Free energy profile for the penetration of polymyxin B1
To elucidate the mechanism by which LPS core sugars prevented penetration of the OM by polymyxin B1, we characterized the free energy profile for the penetration of a single polymyxin B1 molecule from the OM surface to the headgroup region of lipid A through umbrella sampling simulations (Fig. 5). Following translocation through the outer core region (8.0 nm < Z < 8.4 nm), the free energy steadily decreased despite a minor plateau around Z = 8.2 nm before reaching the global energy minimum (ΔG = -6.84 kcal/mol) around the interface between the outer and inner core regions. This explains why most polymyxin molecules remained in the outer core region of the OM in the simulations (Fig. 2). The free energy once again climbed (ΔG = 4.94 kcal/mol) when the polymyxin molecule began to insert into the inner core region (7.74 nm < Z < 8.0 nm), before decreasing to a local energy minimum (ΔG = -4.13 kcal/mol) with further penetration (7.6 nm < Z < 7.74 nm). This energy barrier and local energy minimum prevented the majority of polymyxin molecules from inserting into the inner core region of the OM (Fig. 2). Finally, the free energy sharply increased (ΔG = 5.92 kcal/mol) following the gradual penetration into the lipid A region (7.4 nm < Z < 7.6 nm), which explains the simulation observations that no polymyxin molecule entered the lipid A region of the OM (Fig. 2, Fig. 3)
Fig. 5.

Free energy of polymyxin B1 penetration at a 1:1 peptide/LPS ratio. Free energy profile for polymyxin B1 is shown as a function of Z-coordinates of the outer membrane. The transparent colored shallows show the different regions of the Ra LPS leaflet of the outer membrane. The global minimum of the free energy profile was shifted to zero.
4. Discussion
MDR Gram-negative bacteria have become a major threat to public health due to the limited treatment options available. [1], [2], [3] The seriousness of this situation is exemplified by infections caused by carbapenem-resistant K. pneumoniae which have a mortality rate of approximately 30–40% in the United States and 40–50% in Europe. [27] The dire situation of antibiotic resistance is only predicted to worsen, with antibiotic-resistant infections forecast to kill up to 10 million people each year by 2050 if no effective solution is found. [28] In addition to the cell membrane, Gram-negative bacteria contain an OM that is particularly effective in preventing many compounds from entering the cell and reaching their intracellular targets. [29] Polymyxins target the Gram-negative OM directly and are often the only available therapeutic option for treatment of infections caused by Gram-negative ‘superbugs’. [4]
Very few studies have previously utilized MD simulations to investigate the activity of the polymyxins against Gram-negative organisms. In these studies, the simulated bacterial OM involved either simple phospholipid bilayers [15] or bilayers incorporating the simplest form of lipid A to represent LPS. [6], [11] In the present study, we developed a bacterial OM model incorporating Ra LPS to examine the interaction of polymyxins with the OM during the penetration process. The structure of Ra LPS contained both lipid A and an entire oligosaccharide chain, and thus more accurately reflects the true situation in Gram-negative bacteria. The hydrocarbon tails and negatively charged phosphate groups of lipid A and the oligosaccharide (e.g., carboxyl groups) constitute a permeability barrier of the bacterial OM. As will be discussed further below, we found that the anionic core sugars, especially those of the inner core, severed as an intrinsic barrier to polymyxins. Furthermore, the free energy and cation distribution analysis revealed that calcium ions were essential for the formation of this intrinsic antibiotic-defense mechanism.
Divalent cations are believed to stabilize the bacterial OM. [13], [26], [30] In our study, calcium ions formed a high-density shell within the inner core of LPS (Fig. 4A), mediating bridging interactions between the negatively charged phosphate groups and anionic sugars of adjacent LPS molecules and strengthening the structural integrity of the OM (Fig. S1). These findings are well supported by previous experimental studies. [31], [32] Notably, a major free energy barrier appeared when the polymyxin molecule passed through the LPS inner core region where the calcium ions showed the highest distribution (Fig. 4A and 5). As polymyxins penetrated the inner core, they competitively displaced the calcium ions from the OM (Fig. 4B). These results indicated that cation displacement by polymyxins is energy-unfavorable and the calcium ions in the LPS core sugar regions play a key role in resisting the actions of antimicrobial peptides, including the polymyxins. This agrees with an earlier study that showed that increasing the concentration of calcium ions in solution by 2 μM significantly increased the minimum inhibitory concentrations (MICs, by 2–128 fold) of polymyxins against Pseudomonas species. [33] Furthermore, X-ray and neutron reflectivity experiments showed that calcium removal perturbed the bilayer asymmetry and caused destabilization of the LPS-containing OM. [34] Thus, divalent cations stabilize the OM [8] and provide a level of defense against polymyxins (Fig. 5, Fig. 1S), with their displacement being a prerequisite for initiating membrane disruption and subsequent membrane penetration by polymyxins.
Our results indicate that the core sugar region of LPS functions like an ‘energy trap’. The free energy profile showed two energy minima (ΔG = -6.84 and −4.13 kcal/mol) during polymyxin penetration (Fig. 5), located at the interface between the outer and inner core regions and within the inner core region, respectively. This well explained why most polymyxin molecules only bound to the OM surface rather than inserting into the inner core or lipid A regions (Fig. 2, Fig. 3). Most likely, electrostatic interactions between the anionic sugars and polycationic polymyxin peptides significantly restricted the diffusion of polymyxins, thereby inhibiting the penetration. The most significant OM penetration observed in the MD system containing 96 polymyxin molecules supports their concentration-dependent killing. [35] This indicates that the binding of multiple polymyxin molecules with LPS concertedly disrupts the lateral interaction of the OM, promoting the insertion of other polymyxin molecules (i.e. ‘self-promoted uptake’). [36] It has previously been reported that mutations within the genes responsible for the incorporation of core sugars into LPS attenuated the survival of Burkholderia cenocepacia in vivo. [12] In that study, a mutant strain defective in the expression of both hldA and hldD and which produced LPS consisting of a short lipid A-core oligosaccharide devoid of O-antigen (and thus contained fewer calcium ions than usual in the OM) had increased susceptibility to cationic polymyxin B (>16-fold), melittin (>64-fold) and human neutrophil peptide-1 (>4-fold). [12] Furthermore, our MD results demonstrated that the removal of the core sugars from LPS significantly improved the penetration of polymyxins into the hydrophobic region of the OM (Fig. 2D), which is in excellent agreement with the recent neutron reflectometry studies. [37], [38] Thus, the anionic core sugars of LPS serve as the structural basis of intrinsic resistance of Gram-negative bacteria to environmental toxins, especially polycationic antimicrobials. [14] Indeed, several enzymes for the synthesis and assembly of LPS core sugars are currently being investigated as new antimicrobial targets. [39]
Despite the importance of LPS core sugars in resisting antimicrobial peptides, the lipid A of LPS is very likely the key determinant for bacterial defense. We have recently shown that the free energy barrier for the translocation of polymyxins through the lipid A region of the OM was approximately 68 kcal/mol, [6] much higher than that through LPS core sugar region reported herein (~5 kcal/mol, Fig. 5). This supports the simulation observations that showed the polymyxin molecules could not insert into the lipid A region in all three Re LPS-containing simulation systems (Fig. 2). In nature, bacteria develop polymyxin resistance primarily by modifying the negatively-charged phosphate groups of lipid A, which is controlled by two-component regulatory systems, including PhoPQ and PmrAB. [40] These modifications decrease the specific binding of polymyxins to LPS and inhibit the unique folding of polymyxins within the OM, both of which are required for polymyxin activity. [11] Experimentally, the polymyxin MICs against A. baumannii, P. aeruginosa and K. pneumoniae with lipid A-modified OMs are greatly increased (by 32–256 fold). [41] Collectively, these results indicate that lipid A plays a primary defensive role against antimicrobial peptides, while the core sugars of LPS play a secondary role.
5. Conclusions
In conclusion, we employed coarse-grained MD simulations and umbrella sampling to examine the dynamic behavior of polymyxins interacting with an LPS-containing OM in Gram-negative bacteria. Through extensive thermodynamic analysis, our results suggested that the bridging of the LPS core sugars by calcium ions contributed to the formation of a permeability barrier. Compared to the lipid A-mediated permeability barrier, the core sugar-mediated barrier provided lower-level but broad-spectrum protection against polycationic antimicrobials. Inhibition of the biosynthesis of core sugars or their incorporation into LPS would likely increase the susceptibility of Gram-negative bacteria to cationic antibiotics, providing a new strategy for antibiotic combination therapy. Collectively, these results further our understanding of the distinct roles of different parts of bacterial OM in protection from antimicrobial peptides, including the polymyxins.
CRediT authorship contribution statement
Xukai Jiang: Conceptualization, Formal analysis, Writing - original draft. Yuliang Sun: Data curation, Formal analysis. Kai Yang: Formal analysis. Bing Yuan: Formal analysis. Tony Velkov: Software. Lushan Wang: Software. Jian Li: Conceptualization, Project administration, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI132154). JL is an Australia National Health Medical Research Council (NHMRC) Principal Research Fellow. The simulations were performed on the HPC Cloud Platform (National Key Research and Development Project, 2016YFB0201702) at Shandong University (China) and the supercomputer M3 at eResearch, Monash University (Australia). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.051.
Contributor Information
Xukai Jiang, Email: Xukai.jiang@sdu.edu.cn.
Jian Li, Email: Jian.li@monash.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.MacLean R.C., San Millan A. The evolution of antibiotic resistance. Science. 2019;365(6458):1082–1083. doi: 10.1126/science.aax3879. [DOI] [PubMed] [Google Scholar]
- 2.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 3.Imai Y.u., Meyer K.J., Iinishi A., Favre-Godal Q., Green R., Manuse S. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576(7787):459–464. doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velkov T., Roberts K.D., Thompson P.E., Li J. Polymyxins: a new hope in combating Gram-negative superbugs? Future Med Chem. 2016;8(10):1017–1025. doi: 10.4155/fmc-2016-0091. [DOI] [PubMed] [Google Scholar]
- 5.Li J., Nation R.L., Turnidge J.D., Milne R.W., Coulthard K., Rayner C.R. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 6.Jiang, X., Yang, K., Yuan, B., Han, M., Zhu, Y., Roberts, K. D., Patil, N. A., Li, J., Gong, B. and Hancock, R. E. (2020) Molecular dynamics simulations informed by membrane lipidomics reveal the structure–interaction relationship of polymyxins with the lipid A-based outer membrane of Acinetobacter baumannii. J Antimicrob Chemother 75, 3534-3543. [DOI] [PMC free article] [PubMed]
- 7.Akhoundsadegh N., Belanger C.R., Hancock R.E. Outer membrane interaction kinetics of new polymyxin B analogs in Gram-negative bacilli. Antimicrob Agents Chemother. 2019;63:e00935–19. doi: 10.1128/AAC.00935-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice A., Wereszczynski J. Atomistic scale effects of lipopolysaccharide modifications on bacterial outer membrane defenses. Biophys J. 2018;114(6):1389–1399. doi: 10.1016/j.bpj.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López C.A., Zgurskaya H., Gnanakaran S. Molecular characterization of the outer membrane of Pseudomonas aeruginosa. Biochim Biophys Acta Biomembr. 2020;1862(3):183151. doi: 10.1016/j.bbamem.2019.183151. [DOI] [PubMed] [Google Scholar]
- 10.Vergalli J., Bodrenko I.V., Masi M., Moynié L., Acosta-Gutiérrez S., Naismith J.H. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat Rev Microbiol. 2020;18(3):164–176. doi: 10.1038/s41579-019-0294-2. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X., Yang K., Han M.-L., Yuan B., Li J., Gong B. Outer membranes of polymyxin-resistant Acinetobacter baumannii with phosphoethanolamine-modified lipid a and lipopolysaccharide loss display different atomic-scale interactions with polymyxins. ACS Infect Dis. 2020;6(10):2698–2708. doi: 10.1021/acsinfecdis.0c00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loutet S.A., Flannagan R.S., Kooi C., Sokol P.A., Valvano M.A. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol. 2006;188(6):2073–2080. doi: 10.1128/JB.188.6.2073-2080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berglund N.A., Piggot T.J., Jefferies D., Sessions R.B., Bond P.J., Khalid S. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: a molecular dynamics study. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stead, C. M., Zhao, J., Raetz, C. R. H. and Trent, M. S. (2010) Removal of the outer Kdo from Helicobacter pylori lipopolysaccharide and its impact on the bacterial surface. Mol Microbiol 78, 837-852. [DOI] [PMC free article] [PubMed]
- 15.Khondker A., Dhaliwal A.K., Saem S., Mahmood A., Fradin C., Moran-Mirabal J. Membrane charge and lipid packing determine polymyxin-induced membrane damage. Commun Biol. 2019;2(1) doi: 10.1038/s42003-019-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y., Lu J., Han M.-L., Jiang X., Azad M.A.K., Patil N.A. Polymyxins bind to the cell surface of unculturable Acinetobacter baumannii and cause unique dependent resistance. Adv Sci. 2020;7(15):2000704. doi: 10.1002/advs.v7.1510.1002/advs.202000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., De Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 18.Jefferies D., Hsu P.-C., Khalid S. Through the lipopolysaccharide glass: a potent antimicrobial peptide induces phase changes in membranes. Biochemistry. 2017;56(11):1672–1679. doi: 10.1021/acs.biochem.6b01063. [DOI] [PubMed] [Google Scholar]
- 19.Hsu P.-C., Bruininks B.M.H., Jefferies D., Cesar Telles de Souza P., Lee J., Patel D.S. CHARMM-GUI Martini Maker for modeling and simulation of complex bacterial membranes with lipopolysaccharides. J Comput Chem. 2017;38(27):2354–2363. doi: 10.1002/jcc.24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferies D., Shearer J., Khalid S. Role of o-antigen in response to mechanical stress of the E. coli outer membrane: insights from coarse-grained md simulations. J Phys Chem B. 2019;123(17):3567–3575. doi: 10.1021/acs.jpcb.8b12168. [DOI] [PubMed] [Google Scholar]
- 21.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 22.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 23.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. [Google Scholar]
- 24.Jiang X., Yang K., Yuan B., Gong B., Wan L., Patil N.A. Simulations of octapeptin–outer membrane interactions reveal conformational flexibility is linked to antimicrobial potency. J Biol Chem. 2020;295(47):15902–15912. doi: 10.1074/jbc.RA120.014856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hub J.S., de Groot B.L., van der Spoel D. g_wham: a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J Chem Theory Comput. 2010;6(12):3713–3720. [Google Scholar]
- 26.Santos D.E.S., Pol-Fachin L., Lins R.D., Soares T.A. Polymyxin binding to the bacterial outer membrane reveals cation displacement and increasing membrane curvature in susceptible but not in resistant lipopolysaccharide chemotypes. J Chem Inf Model. 2017;57(9):2181–2193. doi: 10.1021/acs.jcim.7b00271. [DOI] [PubMed] [Google Scholar]
- 27.Lewis K. The science of antibiotic discovery. Cell. 2020;181(1):29–45. doi: 10.1016/j.cell.2020.02.056. [DOI] [PubMed] [Google Scholar]
- 28.O’neill, J. (2014) Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. London: Review on Antimicrobial Resistance.
- 29.Rojas Enrique R., Billings Gabriel, Odermatt Pascal D., Auer George K., Zhu Lillian, Miguel Amanda. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature. 2018;559(7715):617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pink D.A., Truelstrup Hansen L., Gill T.A., Quinn B.E., Jericho M.H., Beveridge T.J. Divalent calcium ions inhibit the penetration of protamine through the polysaccharide brush of the outer membrane of Gram-negative bacteria. Langmuir. 2003;19(21):8852–8858. [Google Scholar]
- 31.Schneck E., Schubert T., Konovalov O.V., Quinn B.E., Gutsmann T., Brandenburg K. Quantitative determination of ion distributions in bacterial lipopolysaccharide membranes by grazing-incidence X-ray fluorescence. Proc Natl Acad Sci U S A. 2010;107(20):9147–9151. doi: 10.1073/pnas.0913737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann Moritz, Schneck Emanuel, Gutsmann Thomas, Brandenburg Klaus, Tanaka Motomu. Bacterial lipopolysaccharides form physically cross-linked, two-dimensional gels in the presence of divalent cations. Soft Matter. 2015;11(30):6037–6044. doi: 10.1039/c5sm01002k. [DOI] [PubMed] [Google Scholar]
- 33.D'Amato Richard F., Thornsberry Clyde, Baker Carolyn N., Kirven Linda A. Effect of calcium and magnesium ions on the susceptibility of Pseudomonas species to tetracycline, gentamicin polymyxin B, and carbenicillin. Antimicrob Agents Chemother. 1975;7(5):596–600. doi: 10.1128/aac.7.5.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clifton Luke A., Skoda Maximilian W.A., Le Brun Anton P., Ciesielski Filip, Kuzmenko Ivan, Holt Stephen A. Effect of divalent cation removal on the structure of gram-negative bacterial outer membrane models. Langmuir. 2015;31(1):404–412. doi: 10.1021/la504407v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nang Sue C., Azad Mohammad A.K., Velkov Tony, Zhou Qi (Tony), Li Jian, Barker Eric. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol Rev. 2021;73(2):679–728. doi: 10.1124/pharmrev.120.000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hancock R.E. Peptide antibiotics. Lancet. 1997;1997(349):418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 37.Paracini Nicolò, Clifton Luke A., Skoda Maximilian W.A., Lakey Jeremy H. Liquid crystalline bacterial outer membranes are critical for antibiotic susceptibility. Proc Natl Acad Sci U S A. 2018;115(32):E7587–E7594. doi: 10.1073/pnas.1803975115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Mei-Ling, Shen Hsin-Hui, Hansford Karl A., Schneider Elena K., Sivanesan Sivashangarie, Roberts Kade D. Investigating the interaction of octapeptin A3 with model bacterial membranes. ACS Infect Dis. 2017;3(8):606–619. doi: 10.1021/acsinfecdis.7b00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cipolla L., Polissi A., Airoldi C., Galliani P., Sperandeo P., Nicotra F. The Kdo biosynthetic pathway toward OM biogenesis as target in antibacterial drug design and development. Curr Drug Discovery Technol. 2009;6:19–33. doi: 10.2174/157016309787581093. [DOI] [PubMed] [Google Scholar]
- 40.Olaitan A.O., Morand S., Rolain J.M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velkov Tony, Gallardo-Godoy Alejandra, Swarbrick James D., Blaskovich Mark. A.T., Elliott Alysha G., Han Meiling. Structure, function, and biosynthetic origin of octapeptin antibiotics active against extensively drug-resistant Gram-negative bacteria. Cell Chem Biol. 2018;25(4):380–391.e5. doi: 10.1016/j.chembiol.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.