FIGURE 2.
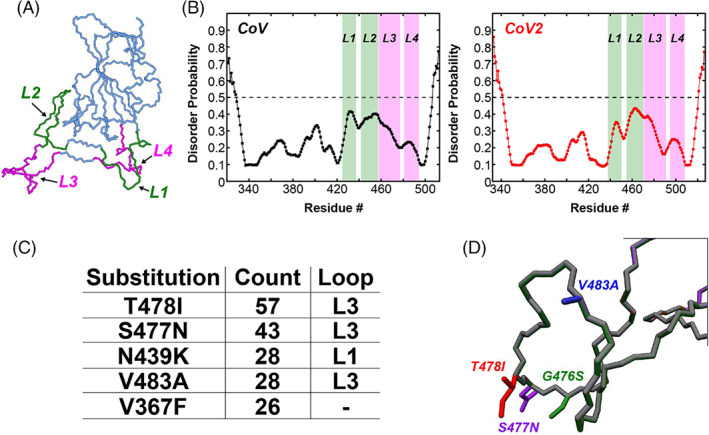
Four loops in the SARS‐CoV‐2 RBD form the binding interface with ACE2 and harbor several single amino acid substitutions identified during the COVID‐19 pandemic. (A) The CoV2‐RBD (PDB 6m0j) showing the four different loops that make up the binding interface (green and pink). Residues 438–450 (CoV2) or residues 425–437 (CoV) make up Loop 1, residues 455–470 (CoV2) or residues 442–457 (CoV) make up Loop 2, residues 471–491 (CoV2) or residues 458–477 (CoV) make up Loop 3, and residues 495–508 (CoV2) or residues 481–494 (CoV) make up Loop 4. (B) Prediction of natively disordered regions using the Protein Disorder prediction System (PrDOS) webserver 12 for CoV‐RBD (left) and CoV2‐RBD (right). PrDOS was used without template‐based prediction and thus reports only on the disorder probability of the local amino acid composition. A prediction false positive rate of 5% was used, and values above the 50% threshold (dotted line) indicate regions of predicted disorder. (C) The five most common mutations of the RBD identified during the first 6 months of the COVID‐19 pandemic, with four of these being located in the loop regions of the binding interface. (D) Depictions of the side‐chains of the mutant residues contained in Loop 3 that are studied in the current work
