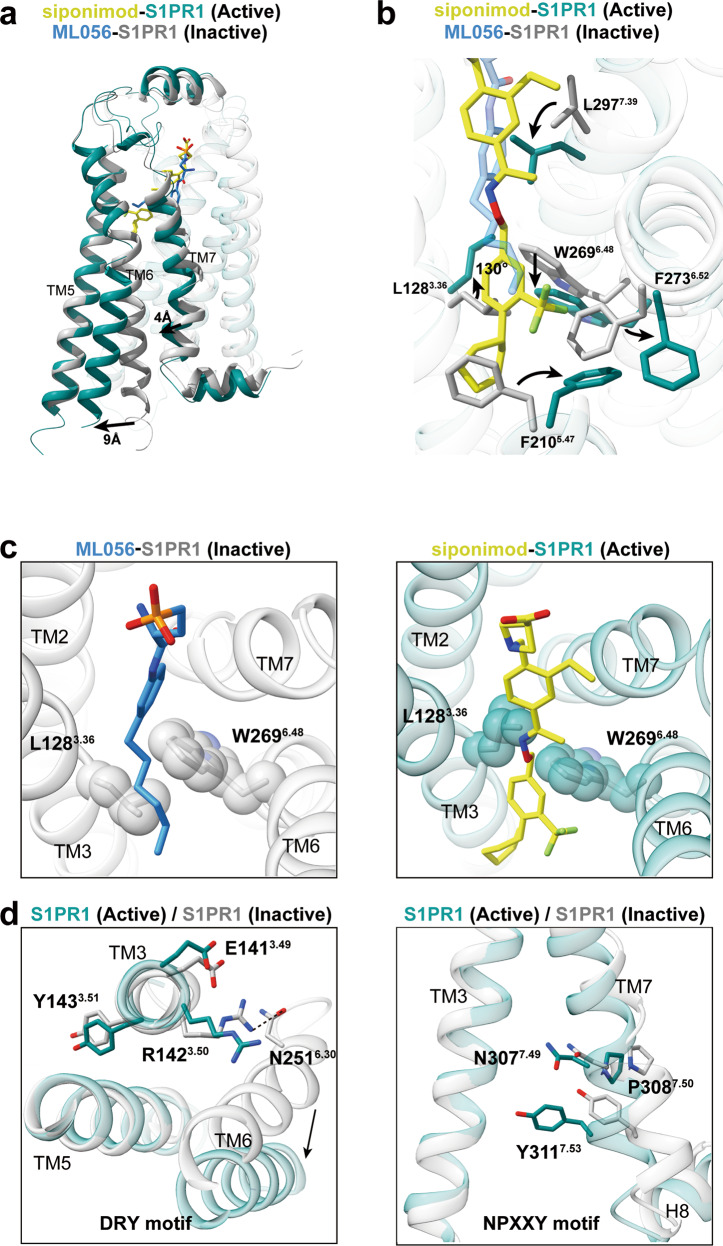
Fig. 4. S1PR1 activation mechanism.
a Superposition of S1PR1 structures in inactive (gray; PDB code: 3V2Y) and Gi-stabilized active states (teal). Notable conformational changes occur at intracellular ends of TM6 and TM7 upon receptor activation. b Detailed comparison of the orthosteric binging pocket of antagonist ML056 (dodger blue)-bound inactive S1PR1 with that of agonist siponimod-bound active S1PR1. In particular, side-chains of the residues L1283.36, F2105.47, W2696.48, F2736.52 and L2977.39 are observed to exhibit a notable displacement upon activation. c The toggle molecular switch L1283.36 and W2696.48 in S1PR1. The left panel displays relative orientation of L1283.36–W2696.4 in inactive S1PR1, whereas the right panel represents movement of L1283.36–W2696.4 when sensing agonist. d The key D/E–R3.50–Y and N–P7.50–xx–Y7.53 motifs important for activation of S1PR1 are observed to display conformational rearrangement in activated receptor.