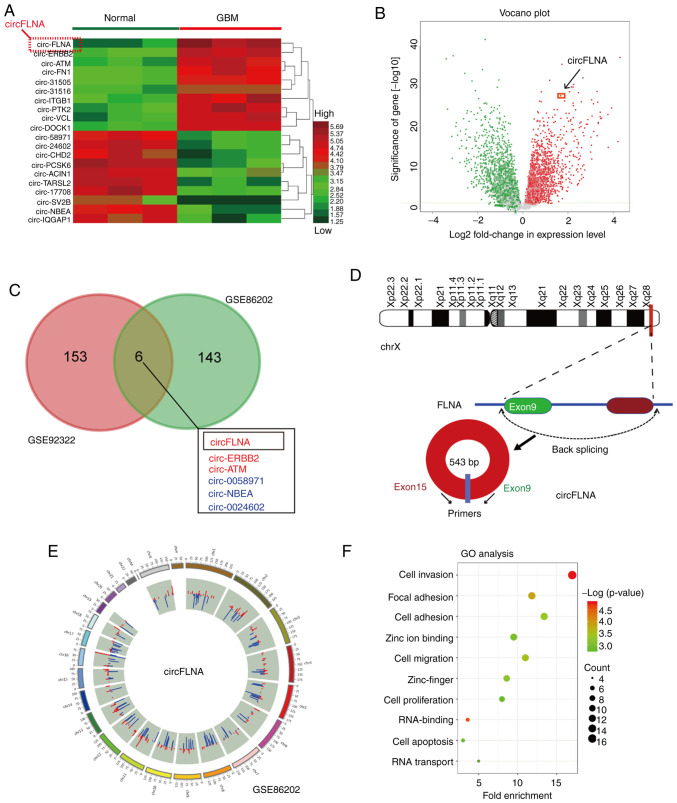
Figure 1.
circFLNA expression levels are upregulated in GBM. (A) Heatmap demonstrating the differentially expressed circRNAs (fold-change) in the GEO dataset, GSE86202. (B) Volcano plot indicated that circFLNA expression levels were increased in the two groups of circRNAs from the GEO database. (C) A total of six significantly differentially expressed circRNAs were identified in the GSE86202 and GSE92322 GEO datasets. (D) Genomic loci of the FLNA gene and formation of circFLNA. The spliced mature sequence length of circFLNA is 543 base pairs in length. (E) Overview of differentially expressed circRNAs and their chromosomal location (using the GSE86202 dataset). The blue bar (downregulated circRNAs) and red bar (upregulated circRNAs) formed the inner circle. (F) Genes positively associated with circFLNA in the GEO datasets (GSE86202) were identified using GO analysis. circRNA, circular RNA; circFLNA, circRNA filamin A; GBM, glioblastoma; GEO, Gene Expression Omnibus; TPM, trans per kilobase of exon model per million; GO, Gene Ontology; Chr, chromosome.