Figure 1. Quantified miRNAs in Serum from Rodents Exposed to Ground Analogs of Spaceflight.
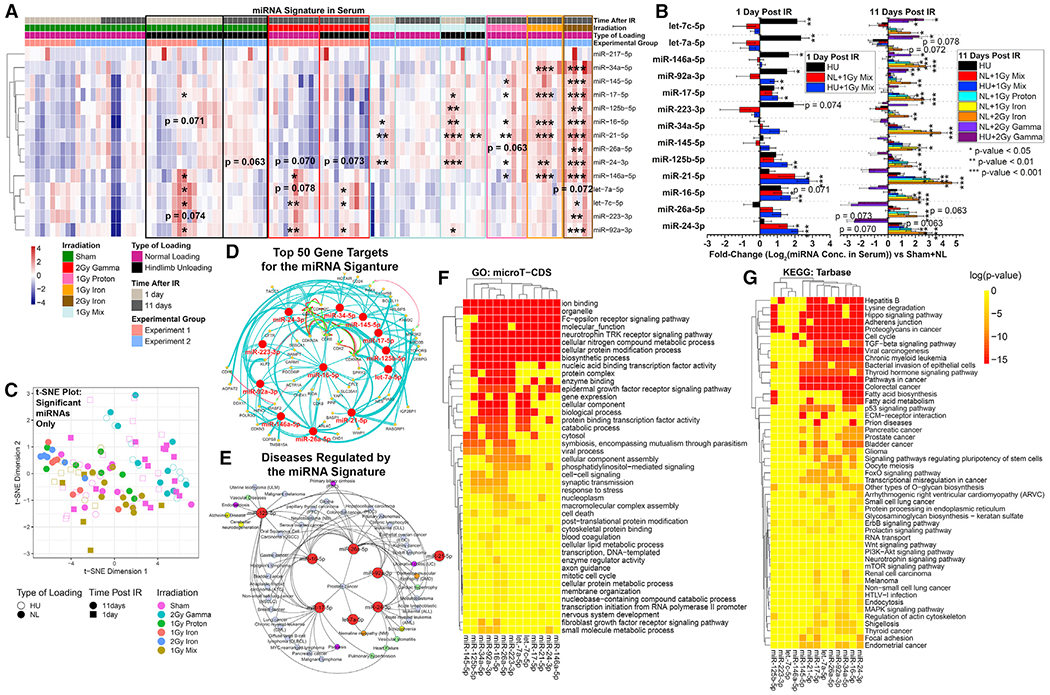
Microgravity and radiation conditions of spaceflight were simulated alone and in combination.
(A) Quantification of miRNAs in serum from rodents exposed to hindlimb unloading (HU) or normal loading (NL) for 3 days before 2 Gy gamma, 1 Gy proton, 1 Gy 600MeV/n 56Fe, 2 Gy 600MeV/n56Fe, or sham irradiation. HU or NL was continued for another 1 or 11 days after irradiation.
(B) Fold changes (log2) of miRNAs compared to NL sham mice. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t test. The error bars represent SEM.
(C) t-Distributed Stochastic Neighbor Embedding (t-SNE) plot showing how the significantly regulated miRNAs are associated with each experimental group. Statistical significance is based on comparing each group with the sham irradiated NL mice.
(D) Top 50 predicted gene targets for all statistically significant miRNAs determined by Cytoscape plugin ClueGo.
(E) The predicted diseases regulated by the miRNA signature determined through miRNet. The blue nodes represent cancer; green, cardiovascular disease; yellow, neurological diseases; orange, muscle degeneration; purple, digestive issues.
(F) Gene Ontology (GO) pathways predicted to be regulated by all statistically significant spaceflight-associated miRNAs determined by the DIANA-microT-CDS (v5.0) algorithm.
(G) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways predicted to be regulated by all statistically significant spaceflight-associated miRNAs.
