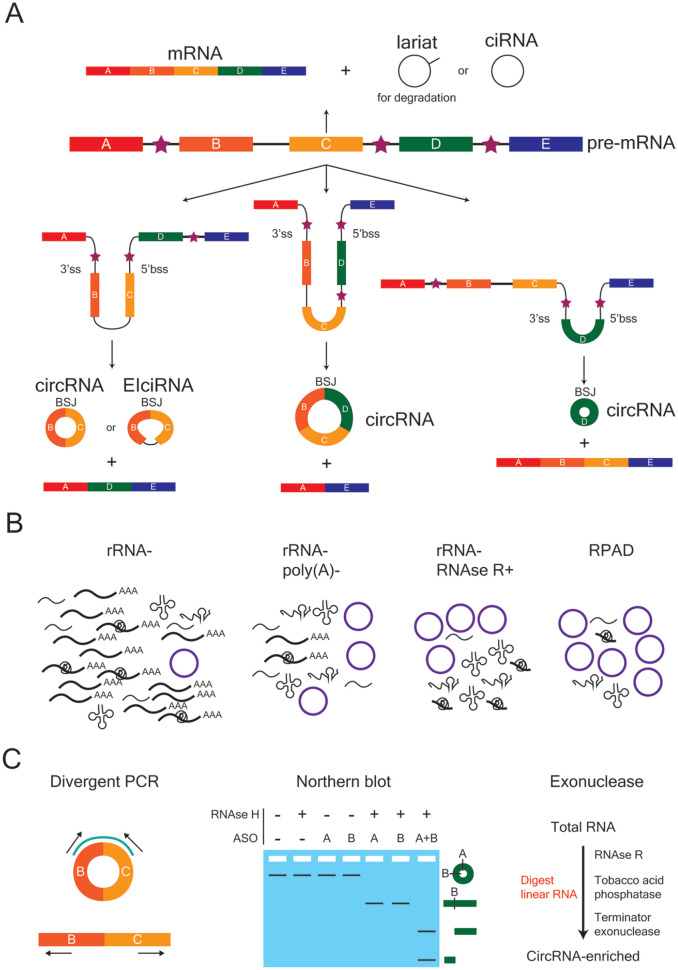
Figure 2.
Biogenesis, enrichment, and validation of circular RNAs (circRNAs). (A) CircRNAs can be exon-only (circRNA), intron-only (ciRNA) or a mixture of both (EIciRNA). The backsplice junction (BSJ) is the site where the 5′ back-splice site (5′bss) and 3′ splice site (3′ss) of a linear strand of RNA are covalently joined. Exons are depicted as rectangular boxes, lettered A to E, whereas introns are represented by the black lines between exons. Stars denote the introns that flank a potential BSJ, which can contain either repetitive or non-repetitive sequences that promote circularization. (B) Several enrichment strategies can be employed to increase the fraction of circRNAs within a given RNA population in order to improve their bioinformatic detection following sequencing. From left to right, the enrichment strategies are rRNA depletion (rRNA−), rRNA depletion and removal of poly(A)+ sequences (rRNA−, poly(A)−), rRNA depletion and RNAse R treatment (rRNA−, RNAse R+), and RNAse R treatment followed by polyadenylation and poly(A)+ depletion (RPAD). (C) Strategies used to verify the circularity of a bioinformatically predicted circRNA. Divergent polymerase chain reaction (PCR) uses outward-facing primers to specifically amplify and detect the region around the BSJ of a predicted circRNA target. These primers are not able to generate a PCR product from the linear RNA of the same gene. A northern blot can be used in combination with anti-sense oligonucleotide (ASO) probes (A, B) and RNAse H treatment to verify circularity. Exonuclease treatment with enzymes that digest linear RNA, followed by divergent PCR, is also commonly used.