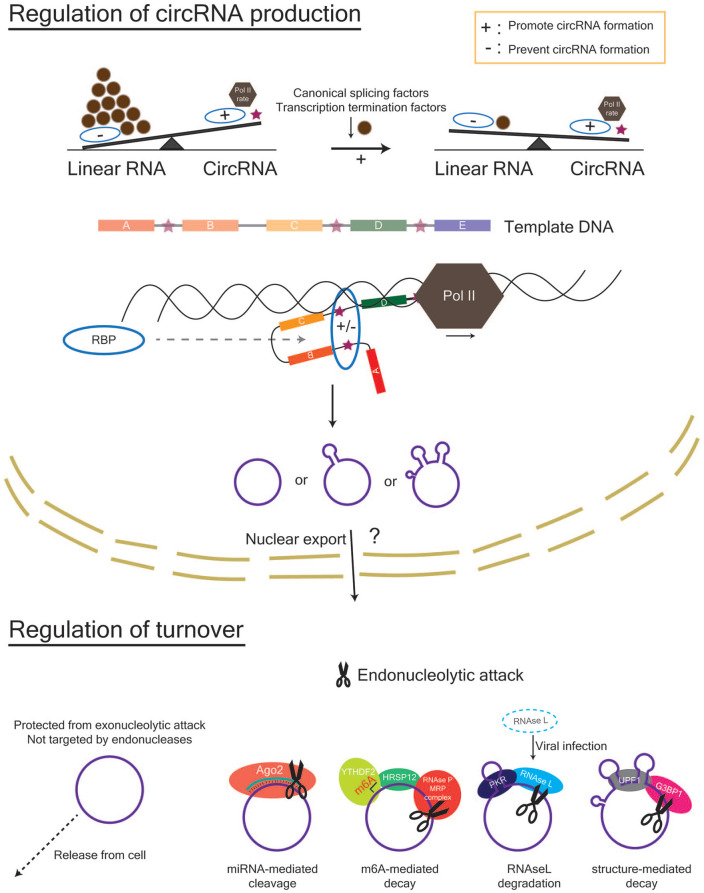
Figure 3.
Regulation of circRNA abundance in the nucleus and cytoplasm. Within the nucleus, the factors that affect the rate and success of circRNA formation include the rate of Pol II transcription, the amount of canonical splicing factors and transcription termination factors, and the presence of RBPs that can bind to sequences within the flanking introns (indicated by stars) of a potential circRNA. Within the cytoplasm, circRNAs can be removed by cellular release via extracellular vesicles or degradation via endonucleolytic attack. Mechanisms for endonucleolytic attack include (1) miRNA-mediated cleavage by Ago2, (2) RNAse L–mediated degradation following viral infection in immune cells, (3) m6A-mediated decay via YTHDF2- HRSP12-RNAseP/MRP interactions, and 4) structure-mediated decay via G3BP1 and UPF1. Ago2, Argonaute 2; G3BP1, G3BP stress granule assembly factor 1; HRSP12, heat-responsive protein 12; m6A, N6-methyladenosine; MRP, multidrug resistance protein; Pol II, polymerase II; RBP, RNA-binding protein; UPF1, UP-Frameshift-1; YTHDF2, YTH N6-methyladenosine RNA binding protein 2.