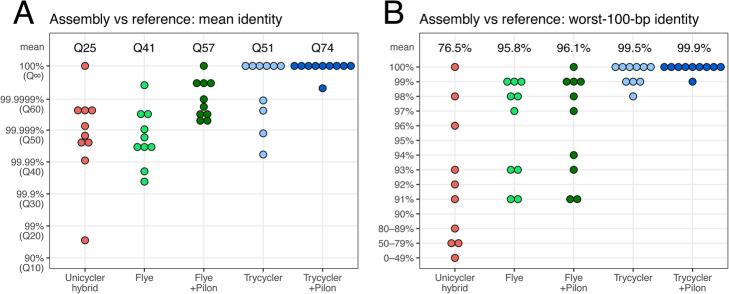
Fig. 2.
Results for the tests using simulated reads. For 10 reference genome sequences, we simulated both short and long reads. The read sets were then assembled with Unicycler (short-read-first hybrid assembly), Flye (long-read-only assembly), Flye+Pilon (long-read-first hybrid assembly), Trycycler (long-read-only assembly), and Trycycler+Pilon (long-read-first hybrid assembly). Each assembled chromosome was aligned back to the reference chromosome to determine the mean assembly identity (A) and the worst identity in a 100-bp sliding window (B). For long-read-only assembly, Trycycler consistently achieved higher accuracy than Flye. Trycycler+Pilon (i.e., using Pilon to polish the Trycycler assembly with short reads) achieved the highest accuracy and did better than alternative hybrid approaches (Unicycler and Flye+Pilon)