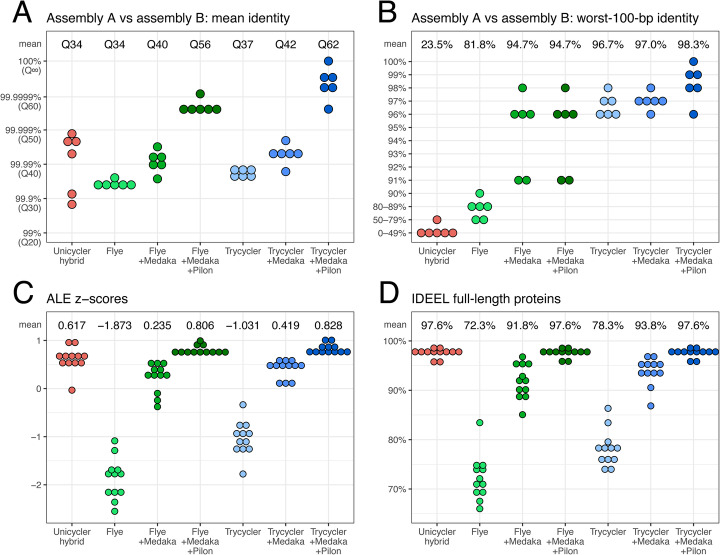
Fig. 3.
Results for the real-read tests. For six genomes, we produced two independent hybrid read sets from the same DNA extraction. The read sets were then assembled with Unicycler (short-read-first hybrid assembly), Flye (long-read-only assembly), Flye+Medaka (long-read-only assembly), Flye+Medaka+Pilon (long-read-first hybrid assembly), Trycycler (long-read-only assembly), Trycycler+Medaka (long-read-only assembly), and Trycycler+Medaka+Pilon (long-read-first hybrid assembly). For each genome and each assembly approach, we aligned the two independently assembled chromosomes to each other to determine the mean assembly identity (A) and the worst identity in a 100-bp sliding window (B). For long-read-only assembly, Trycycler consistently achieved higher accuracy than Flye (both before and after Medaka polishing). Trycycler+Medaka+Pilon achieved the highest accuracy and did better than alternative hybrid approaches (Unicycler and Flye+Medaka+Pilon). We also assessed the accuracy of each of the 12 assembled chromosomes using ALE (C) and IDEEL (D). ALE assigns a likelihood score (transformed into z-scores on a per-genome basis) to each assembly based on its concordance with the Illumina read set. IDEEL identifies the proportion of predicted proteins which are ≥95% the length of their best-matching known protein in a database