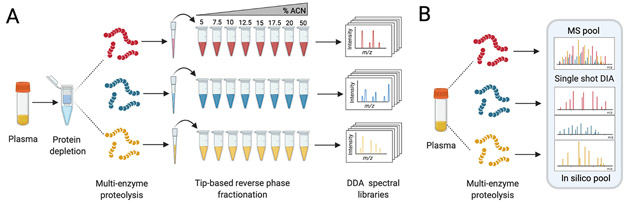
Figure 1. Schematic of the experimental workflow employed.
Graphical representation of the workflow in which plasma is first depleted of high-abundance proteins, digested using either AspN, GluC, or Tryspin, and then individually separated by tip-based high-pH reverse phase fractionation. The resulting fractions are used to generate protease-specific sample libraries by DDA-PASEF(A). Lastly, un-depleted plasma samples digested with the same three proteases are analyzed using DIA-PASEF either individually, or as mixture (1:1:1) the three proteases for a give sample (MS pool). All DIA experiments utilize the aforementioned spectral library (B).