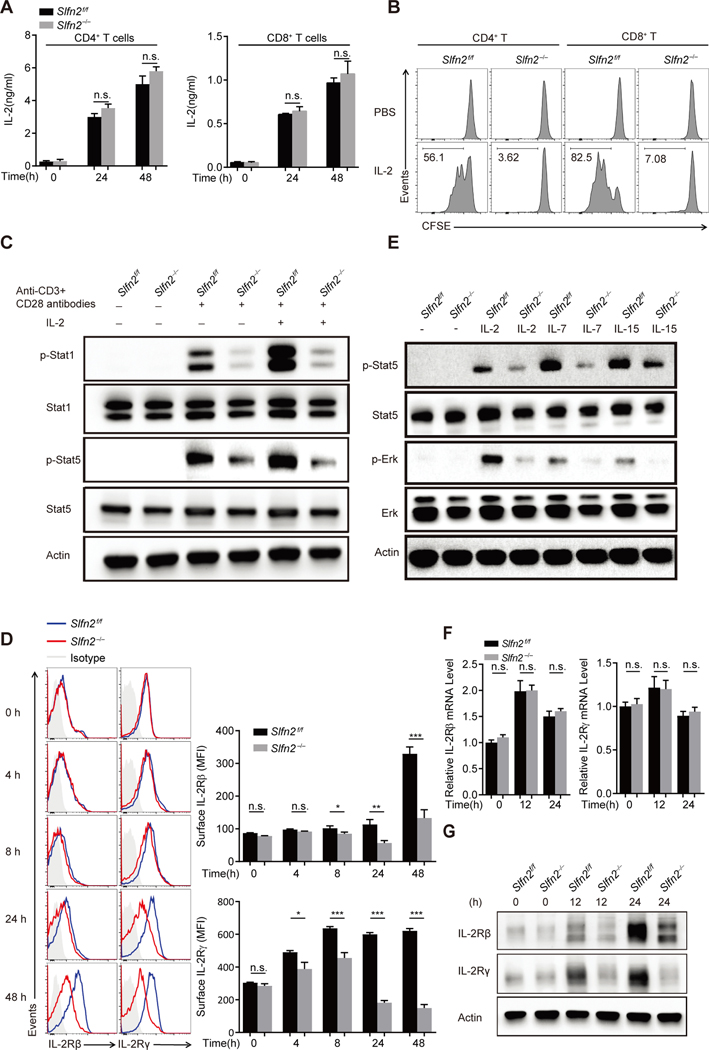
Fig. 2. Abrogation of responses to IL-2 is due to diminished IL-2 receptor expression in SLFN2-deficient T cells.
(A) IL-2 concentration in the culture medium of Slfn2f/f or Slfn2−/− (from CD4-Cre; Slfn2f/f mice) CD4+ and CD8+ T cells left unstimulated or stimulated with anti-CD3/CD28 for 24 hours and 48 hours (n=4 per genotype). (B) Representative flow cytometry analysis of CFSE intensity in Slfn2f/f or Slfn2−/− splenic CD4+ and CD8+ T cells that were pre-activated with anti-CD3/CD28 for 24 hours, then labeled with CFSE, and treated with IL-2 for 48 hours in the absence of anti-CD3/CD28 antibody (n=3 per genotype). (C) Immunoblot analysis of total and phosphorylated Stat1 and Stat5 in Slfn2f/f or Slfn2−/− splenic T cells pre-activated with anti-CD3/CD28 for 24 hours followed by IL-2 stimulation for 30 min with cells pooled from 4–8 mice per genotype. (D) Representative flow cytometry analysis (left) and mean fluorescence intensity (right) of surface expression levels of IL-2Rβ and IL-2Rγ on Slfn2f/f or Slfn2−/− splenic T cells stimulated with anti-CD3/CD28 for the indicated time (n=4 per genotype). Isotype, Slfn2f/f T cells stained with isotype-matched control antibody. (E) Immunoblot analysis of total and phosphorylated Stat5 and Erk in Slfn2f/f or Slfn2−/− splenic T cells pre-activated with anti-CD3/CD28 for 24 hours followed by IL-2, IL-7, or IL-15 stimulation for 30 min, and cells were pooled from 4–8 mice per genotype. (F) qRT-PCR analysis of IL-2Rβ and IL-2Rγ transcripts in total mRNA isolated from Slfn2f/f or Slfn2−/− splenic T cells stimulated with anti-CD3/CD28 for the indicated time (n= 4 per genotype). Data were normalized to β-actin mRNA expression. (G) Immunoblot analysis of IL-2Rβ and IL-2Rγ in Slfn2f/f or Slfn2−/− splenic T cells stimulated with anti-CD3/CD28 for the indicated time with cells pooled from 4–8 mice per genotype. P-values were determined by Student’s t test (n.s., not significant, *P<0.5, **P<0.01, ***P<0.001). Data are representative of two (D) or three (A to C, E to G) independent experiments, and error bars indicate SD.