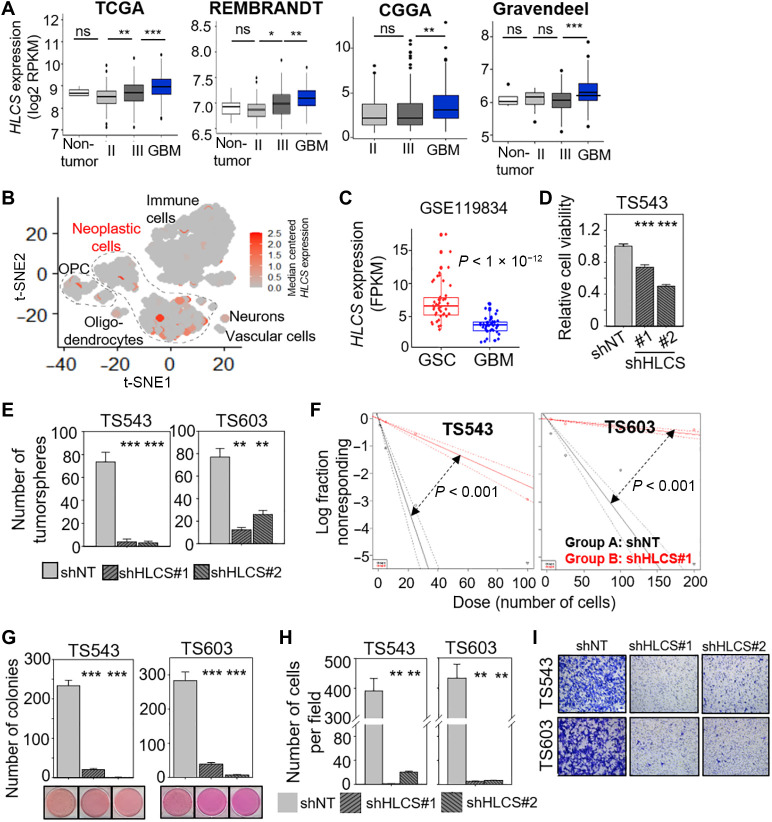
Fig. 6. Genetic disruption of biotin distribution by HLCS depletion impairs GSC proliferation and invasiveness.
(A) Correlative analysis of HLCS mRNA levels with glioma grades in four different glioma patient cohorts. Wilcoxon-Mann-Whitney exact test. *P < 0.05; **P < 0.01; ***P < 1 × 10−4. (B) scRNA-seq data from GBM specimens derived from GSE84465 (31) presented as a t-distributed stochastic neighbor embedding (t-SNE) plot demonstrating expression of HLCS across GBM cells, neurons, oligodendrocyte precursor cells (OPC), oligodendrocytes, and immune cells. Data are presented as median-centered mRNA expression in counts per million (CPM). (C) Correlative analysis of HLCS expression in GSC versus bulk GBM tumors from GSE119834 (45). (D) Cell viability assay of GSC with or without HLCS KD (n = 6) (means ± SD). (E) Tumorsphere formation of GSCs following HLCS KD (n = 6) (means ± SD). (F) Extreme limiting dilution assay of GSCs following HLCS KD (means ± SD). (G) Colony formation of GSC following HLCS KD (n = 4) (means ± SD). Representative images shown below. (H) Transwell migration and invasion assay of GSCs with HLCS KD (n = 3) (means ± SD). (I) Representative images of (H). (D), (E), (G), and (H): **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test.