Pirfenidone inhibits HSC activation and liver fibrosis by targeting glutaredoxin-1 and reducing Smad3 glutathionylation.
Abstract
Activation of the hepatic stellate cells (HSCs) is a key pathogenic event in liver fibrosis. Protein S-glutathionylation (PSSG) of cysteine residues is a distinct form of oxidative response that modifies protein structures and functions. Glutaredoxin-1 (GLRX) reverses PSSG by liberating glutathione (GSH). In this study, we showed that pirfenidone (PFD), an anti-lung fibrosis drug, inhibited HSC activation and liver fibrosis in a GLRX-dependent manner. Glrx depletion exacerbated liver fibrosis, and decreased GLRX and increased PSSG were observed in fibrotic mouse and human livers. In contrast, overexpression of GLRX inhibited PSSG and liver fibrosis. Mechanistically, the inhibition of HSC activation by GLRX may have been accounted for by deglutathionylation of Smad3, which inhibits Smad3 phosphorylation, leading to the suppression of fibrogenic gene expression. Our results have established GLRX as the therapeutic target of PFD and uncovered an important role of PSSG in liver fibrosis. GLRX/PSSG can be both a biomarker and a therapeutic target for liver fibrosis.
INTRODUCTION
Liver fibrosis is often a result of wound-healing response to chronic liver injuries (1). The clinical manifestations of liver fibrosis include excessive deposition of extracellular matrix (ECM) proteins, leading to distortion of the liver architecture and function. Unmanaged liver fibrosis may progress to cirrhosis, which is a key risk factor for portal hypertension, liver failure, and hepatocellular carcinoma (2). There is no U.S. Food and Drug Administration–approved pharmacotherapy for liver fibrosis or cirrhosis (3).
Activation of hepatic stellate cells (HSCs) is a major pathogenic event in liver fibrosis, because activated HSCs are the major source for the production of ECM and profibrogenic cytokines (4). Following persistent liver damage or in vitro culture, quiescent HSCs are activated and transdifferentiate to myofibroblasts with proliferative, contractile, migratory, proinflammatory, and fibrogenic properties (5). Major efforts have been dedicated to better understand the molecular basis of HSC activation with the hope to develop novel strategies to combat liver fibrosis. The canonical transforming growth factor β (TGFβ)–Smad3 pathway plays a key role in the activation of HSCs. TGFβ forms a complex with TGFβ receptor I (TGFBRI) and TGFBRII, leading to the phosphorylation of TGFBRI and creation of a docking site for Smad2/3, predominantly Smad3. Smad anchor for receptor activation (SARA; also called ZFYVE9) captures and presents Smad3 to the binding site of activated TGFBRI, which induces Smad3 activation by phosphorylation (6). The phosphorylation of Smad3 allows the binding to the common mediator Smad4 and increases its nuclear affinity and recruitment of other cofactors for the transcriptional activation of fibrogenic genes.
Another key signal for HSC activation and liver fibrosis is oxidative stress (7). The involvement of oxidative stress is detected in fibrosis with different causative agents concomitant with reduced antioxidant defense (8). During chronic liver injury, oxidative stress–related molecules may act as mediators for HSC activation (9). Compared to the hepatocytes (HEPs), the nonparenchymal cells (NPCs) including HSCs are believed to be more sensitive to oxidants (8). Protein S-glutathionylation (PSSG) is a distinct form of protein oxidation (10). Under oxidative stress, the reactive cysteine residues of reduced proteins are covalently conjugated by glutathione (GSH) to form PSSG. PSSG is an important regulatory mechanism for oxidation to modulate signal transductions by altering the structure and function of target proteins. PSSG is reversible. Glutaredoxin-1 (GLRX), a 12-kDa oxidoreductase, catalyzes protein deglutathionylation and reverses PSSG by liberating GSH from cysteine residues and reestablishing the thiol groups (11). GLRX is highly expressed in the liver, but the cell type specificity of GLRX expression in the liver is unclear (12). It is also unknown whether and how GLRX plays a role in liver fibrosis.
Pirfenidone (PFD; also known as Esbriet) was approved for the treatment of idiopathic pulmonary fibrosis, but without a defined therapeutic target (13). Clinical trials are being conducted, but PFD is yet to be approved for human liver fibrosis (14). PFD has been reported to inhibit HSC activation and fibrosis in rodents but again without a defined molecular target (15–18). It was also suggested that PFD may target other hepatic cell types, such as the HEPs (19).
In this study, we establish GLRX as the therapeutic target of PFD. PFD inhibits HSC activation and liver fibrosis in a GLRX-dependent manner. A forced expression of GLRX was sufficient to inhibit or reverse liver fibrosis, whereas Glrx knockdown or ablation sensitized mice to liver fibrosis. Our study uncovered an indispensable role of protein glutathionylation in HSC activation and liver fibrosis. GLRX is a novel therapeutic target for liver fibrosis.
RESULTS
PFD inhibits HSC activation and liver fibrosis and induces the expression of Glrx in a Stat5-dependent manner
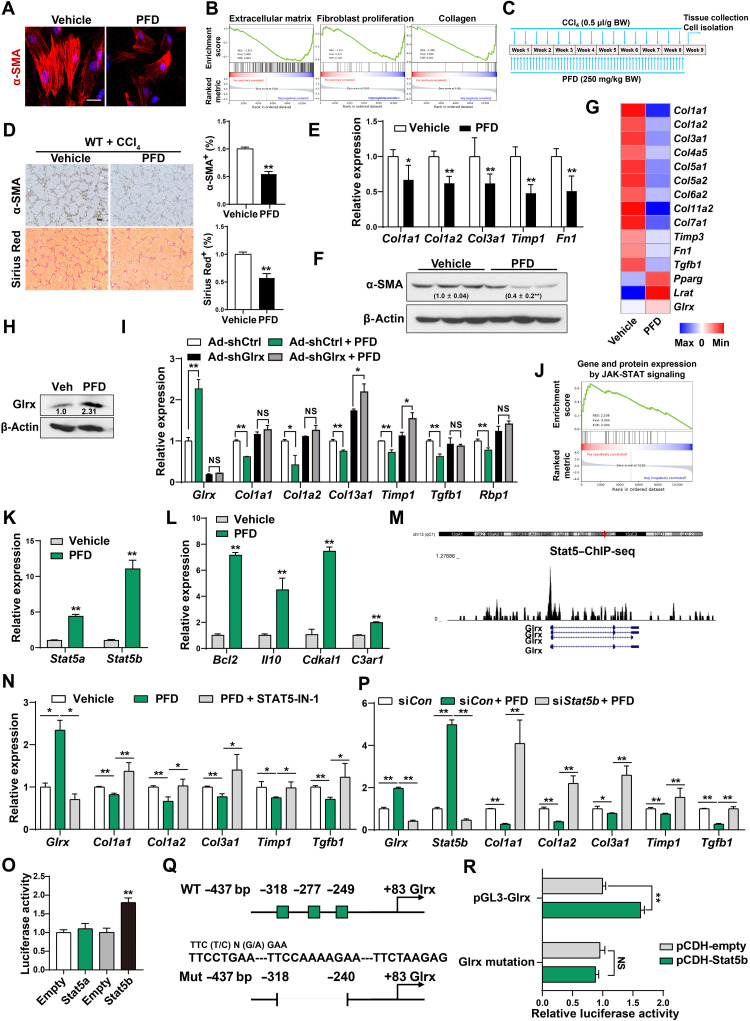
PFD treatment inhibited the culture activation of primary mouse (Fig. 1A) and human HSCs (fig. S1A) as shown by markedly reduced immunostaining of α-SMA (α-smooth muscle actin), consistent with a previous report (16). Gene set enrichment analysis (GSEA) of our RNA sequencing (RNA-seq) data derived from vehicle- and PFD-treated mouse primary HSCs showed substantial down-regulation of fibrogenic pathways (Fig. 1B). Using a regimen of PFD outlined in Fig. 1C, we showed that treatment of wild-type (WT) mice with PFD ameliorated carbon tetrachloride (CCl4)–induced liver fibrosis, as shown by reduced immunostaining of α-SMA and collagen deposition (Fig. 1D), and inhibition of mRNA expression of fibrogenic marker genes (Fig. 1E) and protein expression of α-SMA (Fig. 1F). PFD administration did not affect the serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels in CCl4-treated mice (fig. S1B). Moreover, PFD treatment showed little effect on CCl4- or thioacetamide (TAA)–induced toxicity in primary mouse HEPs as measured by the ALT release assay (fig. S1C). Treatment of WT mice with PFD also inhibited bile duct ligation (BDL)–induced liver fibrosis (fig. S1, D to F).
Fig. 1. PFD inhibits HSC activation and liver fibrosis and induces the expression of Glrx in a Stat5-dependent manner.
(A) Immunofluorescence staining of α-SMA in primary mouse HSCs treated with PFD (1 mM) for 4 days. Scale bar, 50 μm. (B) GSEA of RNA-seq data from PFD-treated primary mouse HSCs. (C to F) Eight-week-old male WT mice were subjected to the CCl4 model and treated with vehicle (n = 4) or PFD (250 mg/kg) (n = 4) by daily gavage (C). α-SMA immunostaining and Sirius Red staining are shown. Scale bar, 200 μm. (D) mRNA expression of fibrogenic genes (E) and protein expression of α-SMA with the quantifications labeled (F). (G) Heat map of HSC gene expression. (H) Protein expression of Glrx. (I) Gene expression in HSCs infected with Ad-shCtrl or Ad-shGlrx and treated with PFD. (J to L) Primary mouse HSCs were treated with PFD (1 mM) for 4 days. GSEA of gene and protein expression by JAK-STAT signaling (J) and mRNA expression of Stat5a, Stat5b (K), and Stat5 downstream genes (L) (n = 3) are shown. (M) ChIP sequencing (ChIP-seq) identified a Stat5-binding peak in mouse Glrx gene promoter. (N) Gene expression in HSCs treated with PFD and STAT5-IN-1 (100 μM) for 4 days (n = 3). (O) Glrx promoter luciferase reporter activity in 293T cells cotransfected with Stat5a or Stat5b plasmid (n = 3). (P) Primary mouse HSCs transfected with siCon or siStat5b were treated with PFD (1 mM) for 4 days (n = 3). (Q and R) WT or deletion mutant Glrx promoter reporter activity in 293T cells cotransfected with Stat5b plasmid (n = 3). Data are means ± SD. *P < 0.05 and **P < 0.01. NS, statistically not significant.
In searching for the downstream effector of PFD, our RNA-seq analysis revealed a significant induction of Glrx in PFD-treated mouse primary HSCs, along with the induction of quiescent HSC marker genes Pparg and Lrat (Fig. 1G). Consistent with the deglutathionylation activity of Glrx, the PSSG level was decreased in PFD-treated mouse liver subjected to the CCl4 model (fig. S1G). The induction of Glrx protein expression in PFD-treated mouse HSCs was confirmed by Western blotting (Fig. 1H). Moreover, the inhibitory effect of PFD on culture activation of mouse primary HSCs was markedly attenuated when the expression of Glrx was knocked down by Ad-shGlrx (Fig. 1I).
In understanding the transcriptional regulation of Glrx by PFD, GSEA of RNA-seq data showed a robust up-regulation of Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling in PFD-treated mouse HSCs (Fig. 1J). The expression of Stat5a and Stat5b (Fig. 1K), phosphorylation of Stat5 (fig. S1H), and Stat5 downstream genes (Fig. 1L and fig. S1I) were induced in PFD-treated HSCs, which led to our hypothesis that the mouse Glrx gene is a transcriptional target of Stat5. Our bioinformatic analysis of chromatin immunoprecipitation (ChIP) sequencing data (20) identified a Stat5-binding peak in the Glrx gene promoter (Fig. 1M). At the functional level, treatment of HSCs with the selective inhibitor STAT5-IN-1 (21) abolished PFD-responsive induction of Glrx and inhibition of fibrogenic genes (Fig. 1N). ChIP assay showed an increased recruitment of Stat5 to the gene promoters of Glrx and several known Stat5 target genes in PFD-treated primary mouse HSCs (fig. S1J). Transfection with Stat5b plasmid, but not Stat5a plasmid, transactivated the Glrx promoter luciferase reporter gene (Fig. 1O), consistent with the greater induction of genes predominantly or specifically regulated by Stat5b in PFD-treated HSCs (fig. S1K). The induction of Glrx and Stat5b and inhibition of fibrogenic gene expression by PFD in siCon-transfected HSCs were abrogated by siStat5b knockdown (Fig. 1P), further supporting that Glrx is a transcriptional target of Stat5 in PFD-treated HSCs. Three Stat5b-binding sites were bioinformatically predicted in the Glrx gene promoter (Fig. 1Q), and deletion of these putative binding sites abrogated the transactivation by Stat5b (Fig. 1R). These results demonstrated that Stat5b mediates the induction of Glrx by PFD.
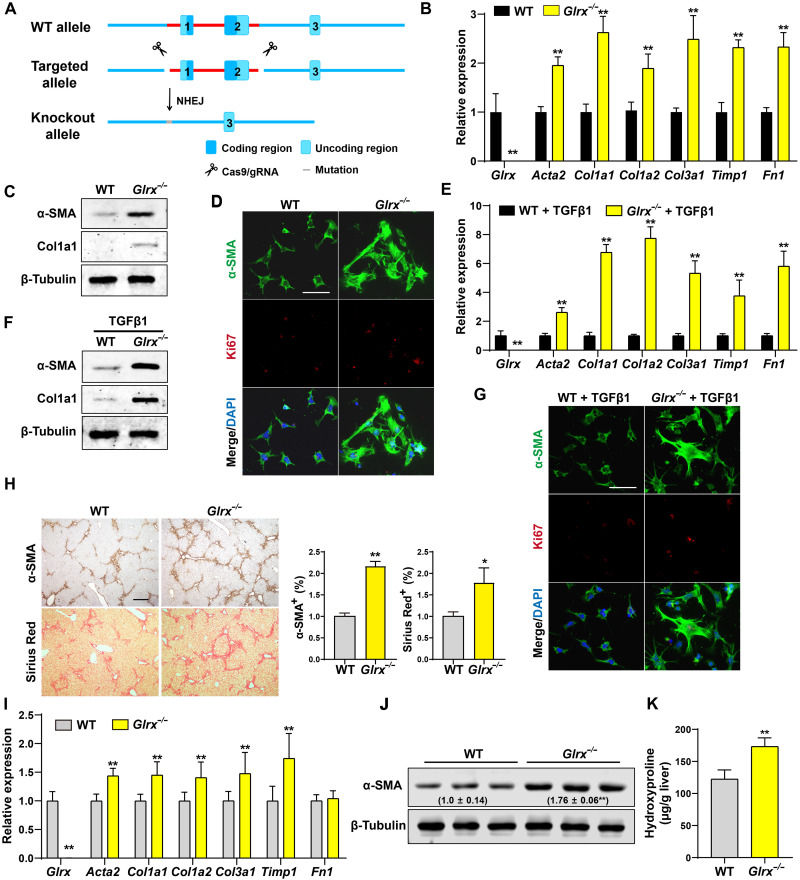
Glrx ablation promotes HSC activation, sensitizes mice to liver fibrosis, and abolishes the anti-fibrotic activity of PFD
To investigate the loss-of-function effect of Glrx in vivo, we generated the whole-body Glrx knockout (Glrx−/−) mice by deleting the two protein-coding exons of the Glrx gene by CRISPR-Cas9–mediated gene targeting (Fig. 2A). The HSCs isolated from Glrx−/− mice showed increased culture activation, as evidenced by the mRNA induction of fibrogenic genes (Fig. 2B), increased protein expression of α-SMA and Col1a1 (Fig. 2C), and increased immunostaining of α-SMA and Ki67 (Fig. 2D). The Glrx−/− HSCs were also more sensitive to TGFβ-stimulated activation, as shown by the mRNA induction of fibrogenic genes (Fig. 2E), increased protein expression of α-SMA and Col1a1 (Fig. 2F), and increased immunostaining of α-SMA and Ki67 (Fig. 2G). In vivo, Glrx−/− mice exhibited increased sensitivity to CCl4-induced liver fibrosis, as evidenced by increased α-SMA immunostaining and Sirius Red staining (Fig. 2H), and mRNA expression of fibrogenic marker genes (Fig. 2I). The increased expression of α-SMA (Fig. 2J) and deposition of collagen (Fig. 2K) were verified by Western blotting and hydroxyproline measurement, respectively. Glrx−/− mice also showed increased sensitivity to BDL-induced liver fibrosis (fig. S2, A to C).
Fig. 2. Glrx ablation promotes HSC activation and sensitizes mice to liver fibrosis.
(A) Schematic representation of the creation of Glrx−/− mice by CRISPR-Cas9 gene targeting. NHEJ, nonhomologous end joining. (B to D) Primary HSCs isolated from WT and Glrx−/− mice were culture-activated for 2 days (n = 3). mRNA expression of Glrx and fibrogenic genes (B), protein expression of α-SMA and Col1a1 (C), and immunofluorescence of α-SMA (green) and Ki67 (red) are shown. Scale bar, 100 μm (D). DAPI, 4′,6-diamidino-2-phenylindole. (E to G) HSCs isolated from WT and Glrx−/− mice were stimulated by TGFβ1 (4 ng/ml) for 1 day (n = 3). mRNA expression of Glrx and fibrogenic genes (E), protein expression of α-SMA and Col1a1 (F), and immunofluorescence of α-SMA (green) and Ki67 (red) are shown. Scale bar, 100 μm (G). (H to K) Eight-week-old male WT and Glrx−/− mice were subjected to the CCl4 model (n = 6). α-SMA immunostaining and Sirius Red staining with the quantifications on the right are shown. Scale bar, 200 μm (H). mRNA expression of Glrx and fibrogenic genes (I), protein expression of α-SMA with the relative quantification values labeled (J), and measurement of liver hydroxyproline level (K). Data are means ± S.D. *P < 0.05 and **P < 0.01, two-tailed Student’s t test.
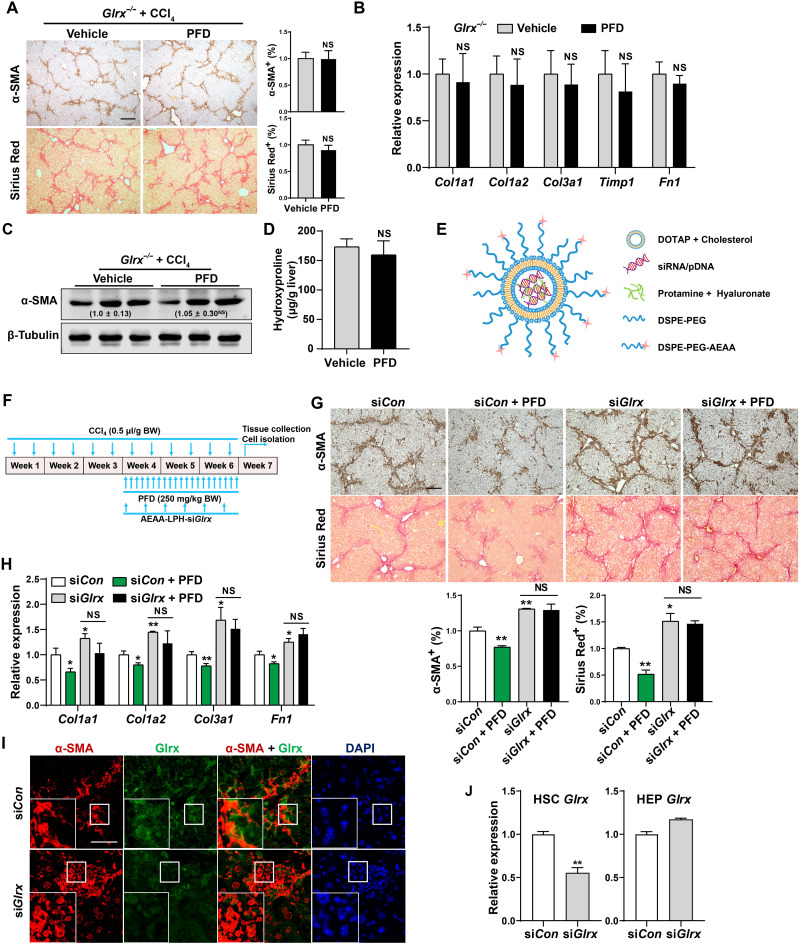
Moreover, the anti-fibrotic activity of PFD was abrogated in Glrx−/− mice, because treatment of Glrx−/− mice with PFD had little effect on CCl4-induced fibrogenesis compared to their vehicle-treated counterparts, as assessed by histology (Fig. 3A), mRNA (Fig. 3B) and protein (Fig. 3C) expression of fibrogenic genes, and hydroxyproline measurement (Fig. 3D).
Fig. 3. Glrx depletion in HSCs abolishes the anti-fibrotic activity of PFD.
(A to D) Eight-week-old male Glrx−/− mice were subjected to the CCl4 model and treated with vehicle or PFD (250 mg/kg body weight) by gavage daily for 8 weeks (n = 6). α-SMA immunostaining and Sirius Red staining with quantifications on the right are shown. Scale bar, 200 μm. (A) mRNA expression of fibrogenic genes (B), protein expression of α-SMA with the relative quantification values labeled (C), and measurement of the liver content of hydroxyproline (D). (E) Schematic representation of the AEAA-conjugated nanoparticles for the delivery of small interfering RNA (siRNA) or plasmids. (F to I) CCl4-treated mice were subjected to nanoparticle regimen and PFD treatment by daily gavages as outlined in (F). α-SMA immunostaining and Sirius Red staining with quantifications at the bottom (n = 4) (scale bar, 200 μm) (G), hepatic mRNA expression of fibrogenic genes (n = 3) (H), and immunofluorescence staining of α-SMA and Glrx in fibrotic livers are shown. Magnified areas of corresponding smaller boxes are shown in the larger boxes. Scale bar, 100 μm (I). (J) Glrx mRNA expression in HSCs and HEPs isolated from mice subjected to a single injection of AEAA-conjugated nanoparticle siCon or siGlrx after a 3-week CCl4 treatment (n = 3). Data are means ± SD. *P < 0.05 and **P < 0.01. Data in (A), (B), and (D) were analyzed by two-tailed Student’s t test. Data in (G), (H), and (J) were analyzed by one-way analysis of variance with the Tukey’s post hoc test.
To determine whether the anti-fibrotic effect of PFD depends on Glrx in HSCs, we used our previously characterized aminoethyl anisamide (AEAA)–conjugated lipid-protamine nanoparticle (22) to deliver siGlrx or shGlrx to knock down Glrx specifically in HSCs in vivo (23). As depicted in Fig. 3E, this AEAA-conjugated nanoparticle preferentially targets activated HSCs, because AEAA is a potent ligand for the Sigma-1 receptor, whose cell surface expression in HSCs is elevated upon activation (24). To demonstrate that this delivery system is efficient and HSC selective, we delivered the enhanced green fluorescent protein (EGFP) plasmid to mice that were subjected to 3-week CCl4 treatment. As shown in fig. S3A, EGFP is mainly expressed in activated HSCs as indicated by the overlap of GFP and α-SMA immunofluorescence. By using a drug regimen outlined in Fig. 3F, PFD treatment also reversed the preestablished liver fibrosis induced by a 3-week CCl4 treatment in siCon nanoparticle–treated mice (Fig. 3, G and H). Nanoparticle delivery of siGlrx exacerbated CCl4-induced progression of liver fibrosis, as evidenced by increased immunostaining of α-SMA and collagen deposition (Fig. 3G) and mRNA expression of fibrogenic genes (Fig. 3H), suggesting that knockdown of Glrx in HSCs was sufficient to sensitize mice to liver fibrosis. Moreover, the anti-fibrotic effects of PFD were abolished in the siGlrx knockdown mice (Fig. 3, G and H). The efficacy and selectivity of the knockdown were verified by decreased Glrx expression in activated HSCs, as shown by immunofluorescence (Fig. 3I), as well as decreased Glrx mRNA expression in primary HSCs, but not HEPs isolated from siGlrx nanoparticle–treated mice (Fig. 3J). Nanoparticle delivery of shGlrx similarly aggravated fibrosis and abolished the anti-fibrotic effects of PFD in the CCl4 model (fig. S3, B to E). These results demonstrated that the HSC Glrx was required for the anti-fibrotic activity of PFD. Meanwhile, primary HEPs whose expression of Glrx was knocked down by Ad-shGlrx or knocked out appeared to be less sensitive to CCl4- or TAA-induced toxicity in vitro as measured by the ALT release assay (fig. S4), suggesting that the loss of Glrx in the HEPs of Glrx−/− mice may not account for the increased sensitivity to fibrosis.
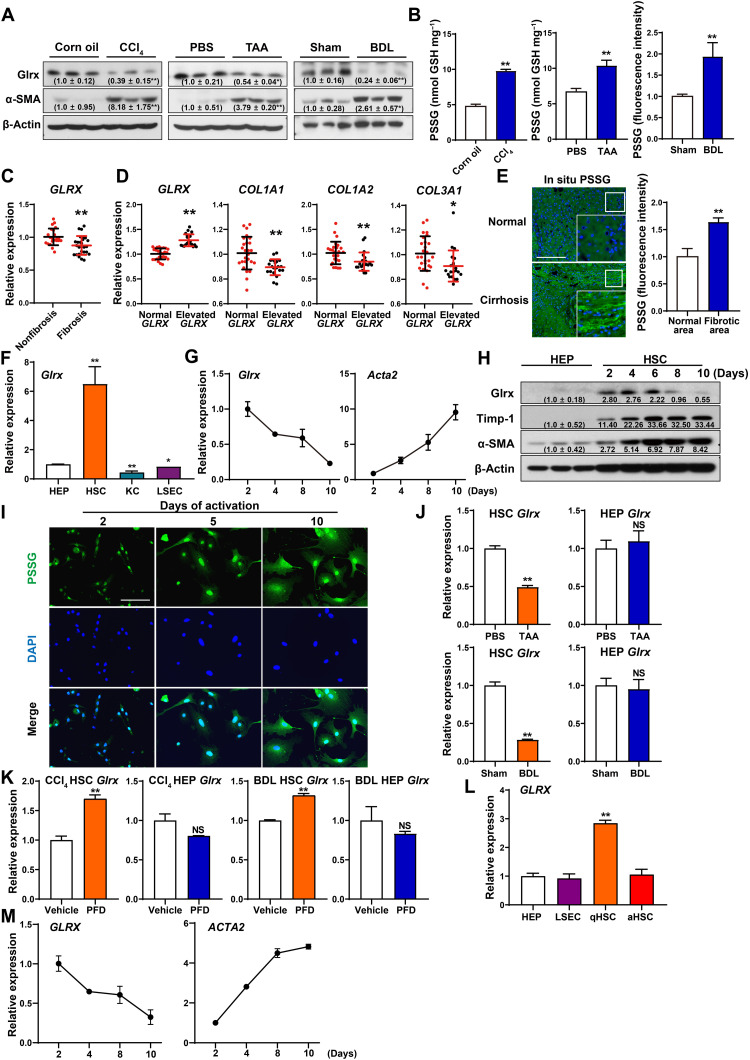
Reduced expression of GLRX and accumulation of PSSG in fibrotic mouse and human livers, and in activated HSCs
Consistent with our observation that down-regulation of Glrx sensitized mice to liver fibrosis, decreased protein expression of Glrx was observed in CCl4-, TAA-, or BDL-induced mouse liver fibrosis, along with the expected up-regulation of α-SMA (Fig. 4A). The success of the fibrosis models was further verified by immunohistochemistry of α-SMA and Sirius Red staining (fig. S5A). The total PSSG levels were increased in all three models as measured by enzymatic recycling assay (for the CCl4 and TAA models) or in situ visualization (for the BDL model) (Fig. 4B), consistent with the notion that GLRX is a deglutathionylation enzyme that reverses PSSG by liberating GSH from cysteine residues (12).
Fig. 4. Reduced expression of GLRX and accumulation of PSSG in fibrotic mouse and human livers, and in activated HSCs.
(A and B) Hepatic protein expression of Glrx and α-SMA (A) and PSSG levels (B) in mice subjected to the CCl4, TAA, or BDL model (n = 3 per group). (C and D) Bioinformatic analyses of GSE15235 derived from patients with fibrotic or non-fibrotic biliary atresia. Hepatic mRNA expression of GLRX (C) and COL1A1, COL1A2, and COL3A1 (D) in patients with normal or elevated GLRX are shown. (E) In situ visualization of PSSG on sections of healthy or fibrotic human livers. Scale bar, 200 μm. (F) Glrx expression in mouse HEPs, HSCs, KCs, and LSECs (n = 3). (G) Glrx and Acta2 expression in culture-activated primary mouse HSCs (n = 3). (H) Protein expression in primary mouse HEPs and culture-activated HSCs. (I) In situ visualization of PSSG in primary mouse HSCs. Scale bar, 100 μm. (J) mRNA expression of Glrx in HSCs and HEPs isolated from mice treated with TAA for three doses or BDL for 2 weeks (n = 3). (K) Glrx expression in HSCs and HEPs isolated from PFD-treated mice subjected to CCl4 or BDL model (n = 3). (L) mRNA expression of GLRX in primary human HEPs, LSECs, quiescent HSCs, and culture-activated HSCs (n = 3). (M) GLRX and ACTA2 expression in culture-activated primary human HSCs (n = 3). Data are means ± SD. *P < 0.05 and **P < 0.01.
The down-regulation of hepatic GLRX mRNA expression was also observed in fibrotic biliary atresia patients compared to their nonfibrotic counterparts [Gene Expression Omnibus (GEO) dataset GSE15235] (Fig. 4C). Analysis of publicly deposited microarray results showed that the expression of GLRX was inversely correlated with the expression of fibrogenic genes (Fig. 4D). Inverse correlations between the expression of GLRX and the fibrogenic marker genes were observed in a TCGA (The Cancer Genome Atlas) cohort of livers that included both liver tumors and normal livers (fig. S5B). Up-regulation of PSSG level was observed in the bridging fibrosis area of cirrhotic human liver, as visualized by in situ PSSG (Fig. 4E).
Liver is an organ of multiple cell types. We then isolated HSCs, HEPs, Kupffer cells (KCs), and liver sinusoidal endothelial cells (LSECs) from the same mice and found that the expression of Glrx was the highest in quiescent HSCs, nearly six times of HEPs (Fig. 4F). The mRNA expression of Glrx in primary mouse HSCs progressively decreased with the onset of culture activation (Fig. 4G and fig. S5C). Western blotting also showed that the Glrx protein expression in primary mouse HSCs was markedly higher than in HEPs, and the HSC expression of Glrx was decreased by culture activation in a time-dependent manner (Fig. 4H). Activated HSCs exhibited a higher level of PSSG (Fig. 4I), consistent with their decreased Glrx expression. The fibrosis-responsive down-regulation of Glrx in HSCs was also confirmed in vivo, as the primary HSCs, but not HEPs isolated from TAA- or BDL-treated mice, showed a decreased expression of Glrx (Fig. 4J). Treatment with PFD restored the expression of Glrx in HSCs without affecting the expression of Glrx in HEPs in CCl4- or BDL-treated livers (Fig. 4K).
In primary human liver cells isolated from the same donor, the mRNA expression of GLRX in quiescent HSCs was approximately three times of HEPs and LSECs (Fig. 4L). The expression of GLRX in primary human HSCs also progressively declined with the onset of culture activation (Fig. 4M and fig. S5D).
Overexpression of GLRX, but not its enzyme-dead mutant, inhibits HSC activation and liver fibrosis in vitro and in vivo
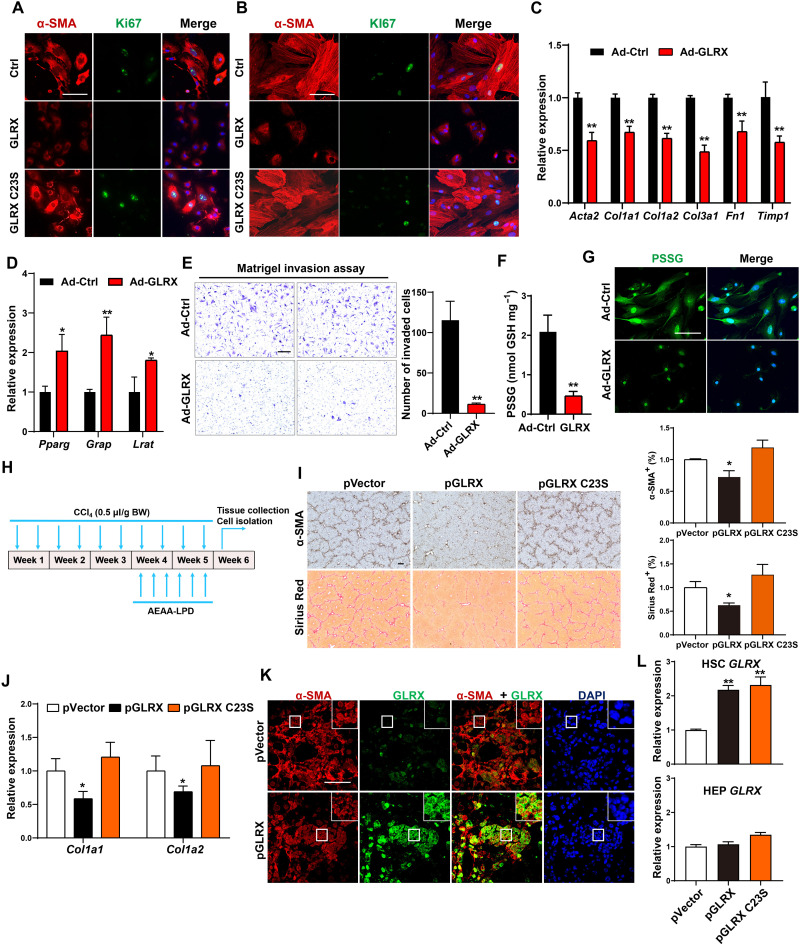
To determine whether overexpression of GLRX can inhibit HSC activation, we infected primary mouse and human HSCs with Ad-GLRX or Ad-GLRX C23S expressing the cysteine-23 to serine mutant GLRX that lacks the oxidoreductase activity (10, 25). Immunofluorescence showed a comparable expression of WT and mutant GLRX (fig. S6A). Infection of mouse HSCs with Ad-GLRX, but not Ad-GLRX C23S, inhibited α-SMA and Ki67 expression (Fig. 5A). A similar pattern of GLRX effect and the lack of GLRX C23S effect on the expression of α-SMA and Ki67 was observed in human HSCs (Fig. 5B).
Fig. 5. Overexpression of GLRX, but not its enzyme-dead mutant, inhibits HSC activation and liver fibrosis.
(A and B) Immunofluorescence staining of α-SMA (red) and Ki67 (green) in primary mouse (A) and human (B) HSCs infected with Ad-GLRX or Ad-GLRX C23S. Nuclei (blue) and DAPI. Scale bar, 100 μm. (C to G) Primary mouse HSCs were infected with Ad-GLRX or Ad-Ctrl 1 day after isolation and culture-activated for an additional 4 days. mRNA expression of activated (C) and quiescent (D) HSC marker genes, Matrigel invasion assay (E), PSSG level detected by enzymatic recycling assay (F), and in situ visualization of PSSG (G) (n = 3) are shown. (H to L) Mice were subjected to the nanoparticle regimen in the CCl4 model as outlined in (H). α-SMA immunostaining and Sirius Red staining with quantifications on the right (n = 4) (scale bar, 200 μm) (I), hepatic mRNA expression of Col1a1 and Col1a2 (n = 4) (J), and immunofluorescence staining of α-SMA and GLRX in fibrotic livers are shown. Magnified areas of corresponding smaller boxes are shown in the larger boxes. Scale bar, 100 μm (K). GLRX mRNA expression in primary HSCs and HEPs isolated at the end of experiments (n = 3) (L). Data are means ± SD. *P < 0.05 and **P < 0.01. Data in (C) to (F) were analyzed by two-tailed Student’s t test. Data in (I), (J), and (L) were analyzed by one-way analysis of variance with the Tukey’s post hoc test.
The inhibition of culture activation of mouse HSCs by Ad-GLRX was further confirmed by decreased fibrogenic gene expression (Fig. 5C), increased quiescent marker gene expression (Fig. 5D), and decreased HSC invasiveness as shown by Matrigel invasion assay (Fig. 5E). As expected, GLRX overexpression decreased the PSSG level as shown by both enzymatic recycling assay (Fig. 5F) and in situ visualization (Fig. 5G). Adenoviral overexpression of GLRX had little effect on HSC death, as shown by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (fig. S6B).
To determine whether overexpression of GLRX can reverse preexisting HSC activation, primary mouse HSCs were culture-activated for 3 days with the activation verified by the induction of fibrogenic genes (fig. S6C). The cells were then infected with Ad-Ctrl or Ad-GLRX and cultured for three additional days. As shown in fig. S6D, overexpression of GLRX inhibited the fibrogenic progression in preactivated HSCs. Adenoviral expression of GLRX also inhibited the fibrogenic program in preactivated passage 5 human HSCs (fig. S6E).
To evaluate the anti-fibrotic effect of GLRX overexpression in vivo, we used the same AEAA-conjugated nanoparticle to deliver GLRX or GLRX C23S plasmid to HSCs. The therapeutic effect of GLRX nanoparticle was tested in preestablished liver fibrosis as outlined in Fig. 5H. Following a 3-week initiation of liver fibrosis by CCl4, the nanoparticles were given by tail vein injections three times a week for 2 weeks, while the CCl4 treatment continued, and the mice were sacrificed 3 days after the last CCl4 and nanoparticle injections. Compared to the empty vector control, delivery of the GLRX plasmid, but not the GLRX C23S plasmid, attenuated the fibrotic progression, as shown by decreased immunostaining of α-SMA and Sirius Red staining of collagens (Fig. 5I), and reduced mRNA expression of fibrogenic marker genes (Fig. 5J). The induction of GLRX in activated HSCs was verified by the overlap of GLRX and α-SMA immunofluorescence using confocal microscopy (Fig. 5K). The mutually exclusive staining of GLRX and HEP nuclear factor 4α (HNF4α; the HEP marker) indicated the minimal off-target event (fig. S6F). The efficient GLRX expression in HSCs was further supported by coexpression of GLRX and Col1a1 by RNA in situ hybridization (fig. S6G). Isolation of primary cells at the end of the experiments showed a comparable induction of GLRX or GLRX C23S in HSCs, but not in HEPs (Fig. 5L). Consistent with our observations in primary HSCs infected with Ad-GLRX, forced expression of GLRX in activated HSCs by nanoparticle delivery decreased PSSG levels in CCl4-treated mouse livers (fig. S6H).
Smad3 is a novel GLRX substrate, and glutathionylation of Smad3 is indispensable for its transactivation of fibrogenic genes
TGFβ is a well-established profibrogenic cytokine, so we went on to determine whether GLRX can inhibit TGFβ-stimulated HSC activation. Treatment of primary mouse HSCs with TGFβ inhibited the expression of Glrx (fig. S7A). TGFβ stimulation resulted in a robust induction of fibrogenic marker genes as expected, but the induction was attenuated when mouse HSCs were infected with Ad-GLRX (Fig. 6A). In contrast, adenoviral short hairpin RNA (shRNA) knockdown of Glrx sensitized mouse HSCs to TGFβ-stimulated HSC activation (fig. S7B). These results were consistent with our RNA-seq results, in which the TGFβ and CO-SMAD binding signaling pathways were inhibited in Ad-GLRX–infected HSCs, along with the suppression of other pathways indicative of HSC activation (fig. S7C).
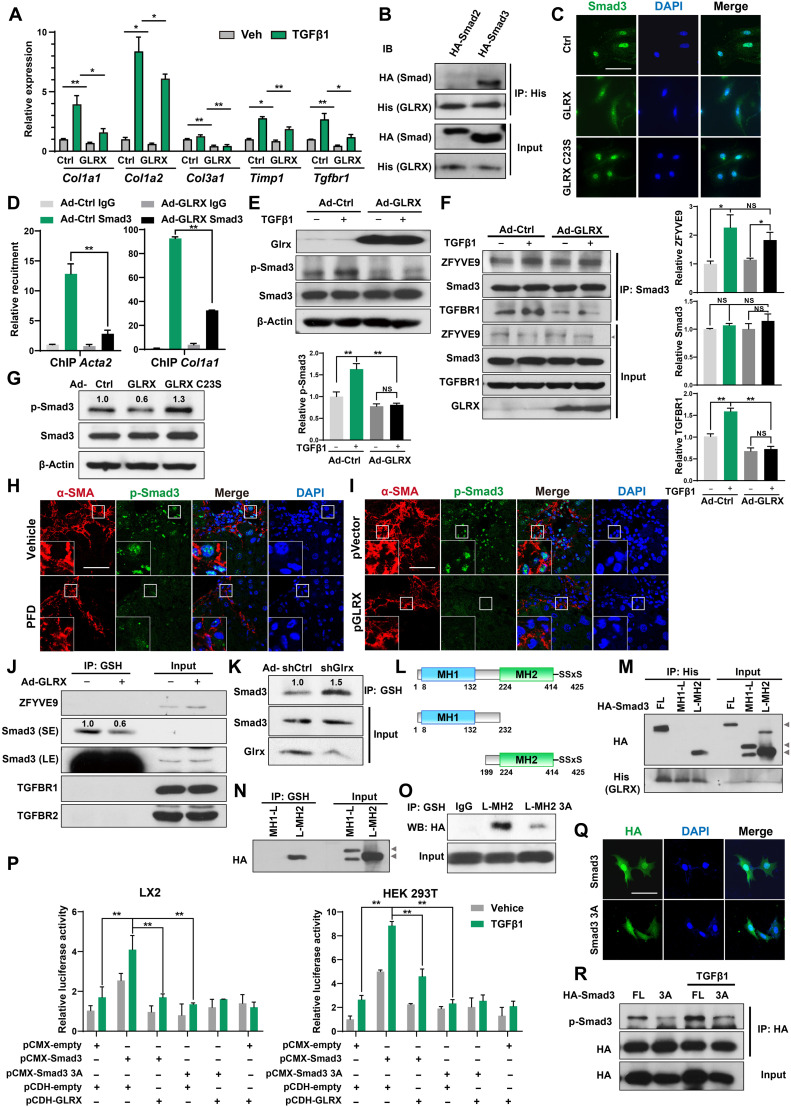
Fig. 6. Smad3 is a GLRX substrate, and glutathionylation of Smad3 is indispensable for its transactivation of fibrogenic genes.
(A) Gene expression in mouse HSCs infected with Ad-GLRX and treated with TGFβ1 (4 ng/ml) for 24 hours. (B) Protein interactions were assessed by co-IP in LX2 cells transfected with HA-Smad2 or HA-Smad3, and His-GLRX plasmids. (C) Immunofluorescence assessment of Smad3 in mouse HSCs infected with Ad-GLRX or Ad-GLRX C23S. Scale bar, 100 μm. (D) ChIP assessment of the recruitment of Smad3 onto the Acta2 and Col1a1 gene promoters in mouse HSCs. (E) Phosphorylation of Smad3 in HSCs assessed by Western blotting. (F) Protein interactions were assessed by co-IP in LX2 cells treated with TGFβ1 (5 ng/ml) for 24 hours. (G) Phosphorylation of Smad3 in HSCs assessed by Western blotting. (H and I) Immunostaining of hepatic α-SMA and phosphorylated Smad3 in CCl4-treated mice subjected to PFD (H) or GLRX nanoparticle (I) treatment. Scale bar, 100 μm. (J and K) PSSG in primary mouse HSCs infected with Ad-GLRX (J) or Ad-shGlrx (K). SE, short exposure; LE, long exposure. (L) Illustration of WT and deletion mutant Smad3. (M and N) LX2 cells infected with Ad-GLRX were transfected with HA-Smad3 plasmids. FL, full-length. Protein interaction between Smad3 and GLRX (M), and Smad3-SSG (N) are shown. (O) Smad3-SSG in LX2 cells transfected with HA-L-MH2 or HA-L-MH2 3A plasmids. (P) SBE-Luc reporter activity in LX2 and 293T cells cotransfected with Smad3 and GLRX plasmids (n = 3). (Q and R) LX2 cells transfected with HA-tagged Smad3 or Smad3 3A were stimulated by TGFβ1 (5 ng/ml) for 24 hours. Nuclear translocation (Q) and phosphorylation (R) of Smad3 are shown. Data are means ± SD. *P < 0.05 and **P < 0.01.
Smad proteins are key effectors of TGFβ that directly transactivate fibrogenic genes. Coimmunoprecipitation (co-IP) assay showed that GLRX can interact with Smad3, but not Smad2, in LX2 cells cotransfected with GLRX and individual Smads (Fig. 6B). At the functional level, the nuclear translocation of Smad3 in activated primary mouse HSCs was inhibited in cells infected with Ad-GLRX but not GLRX C23S (Fig. 6C). Moreover, overexpression of GLRX inhibited the recruitment of Smad3 onto the fibrogenic gene promoters as shown by ChIP assay (Fig. 6D). Smad3 phosphorylation (Ser423/425) is essential for its nuclear translocation and transcriptional activity (6). Overexpression of GLRX in primary mouse HSCs (Fig. 6E) or LX2 cells (fig. S7D) attenuated the basal and TGFβ-stimulated Smad3 phosphorylation.
To determine the molecular events associated with the suppression of Smad3 phosphorylation by GLRX, the expressions and their interactions with Smad3 of several proteins that can directly affect the phosphorylation of Smad3 were assessed by co-IP in LX2 cells. The expression of SARA (ZFYVE9) (26), Smad3, and TGFBRI was not affected by GLRX overexpression (Fig. 6F). Overexpression of GLRX suppressed the TGFβ-stimulated interaction between Smad3 and TGFBRI, without affecting the Smad3-ZFYVE9 interaction (Fig. 6F). The interaction between Smad3 and TGFBRI is responsible for the docking and phosphorylation of Smads (27), suggesting that GLRX may have attenuated Smad3 phosphorylation by sequestration of Smad3 from TGFBRI. Consistent with its lack of inhibitory effect on HSC activation, GLRX C23S failed to inhibit culture activation–induced phosphorylation of Smad3 in primary mouse HSCs (Fig. 6G). The expression of Smad3, Tgfbr1, and Zfyve9 and phosphorylation of Smad3 in HSCs of PFD- and GLRX nanoparticle–treated fibrotic mouse livers were measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and immunofluorescence, respectively. As shown in fig. S7E, PFD or GLRX nanoparticle had little effect on Smad3 and Zfyve9 expression but modestly decreased Tgfbr1 expression in HSCs isolated from CCl4-treated livers. The fibrosis-responsive nuclear accumulation of phosphorylated Smad3 in HSCs was inhibited by PFD (Fig. 6H) or AEAA-GLRX nanoparticle in mice subjected to the CCl4 model (Fig. 6I).
Having shown that GLRX interacted with and inhibited the transcriptional activity of Smad3 and knowing that the enzymatic activity of GLRX is required for its anti-fibrotic activity, we speculated that Smad3 is a deglutathionylation substrate of GLRX. We showed that Smad3 is a target for PSSG. An anti-GSH antibody was used to immunoprecipitate S-glutathionylated proteins in primary mouse HSCs before subjecting them to Western blotting. Among proteins involved in the TGFβ-Smad3 pathway, only Smad3, but not ZFYVE9, TGFBRI, or TGFBRII, exhibited appreciable glutathionylation, and GLRX overexpression decreased the glutathionylation of Smad3 (Fig. 6J). In contrast, knockdown of Glrx increased the glutathionylation of Smad3 (Fig. 6K). Smad3 contains Mad homolog domain 1 (MH1), linker region (L), and MH2 as outlined in Fig. 6L. Our deletion analysis showed that GLRX interacted with MH2, but not MH1 or the linker region (Fig. 6M). Consistent with its binding to GLRX, the MH2 domain was also the target of PSSG (Fig. 6N). There are 10 cysteines in the MH2 domain (fig. S7F). Mutations of individual cysteines to alanines showed that the glutathionylation of L-MH2 C332A, C338A, and C370A was noticeably decreased compared to L-MH2 WT (fig. S7G). We then constructed a triple mutant L-MH2 3A that contained the mutations of C332, C338, and C370 to alanines. The glutathionylation of L-MH2 3A was markedly decreased compared to L-MH2 (Fig. 6O). To determine the functional consequence of the loss of Smad3 glutathionylation, we mutated the same three cysteines in the context of full-length Smad3 (Smad3 3A). The transcriptional activity of Smad3 3A was compared to WT Smad3 in LX2 and 293T cells cotransfected with the Smad3-responsive luciferase reporter SBE-Luc in the absence or presence of GLRX cotransfection or TGFβ1 (5 ng/ml) stimulation. Transfection of WT Smad3 increased basal and TGFβ-stimulated reporter activity as expected, and both effects were abolished by GLRX cotransfection (Fig. 6P). In contrast, transfection of Smad3 3A failed to increase the basal or TGFβ-stimulated reporter activity regardless of the GLRX cotransfection (Fig. 6P). The nuclear translocation of Smad3 3A stimulated by TGFβ was also reduced (Fig. 6Q). Compared to Smad3 WT, transfection of Smad3 3A also showed reduced basal and TGFβ-stimulated phosphorylation (Fig. 6R) and decreased basal and TGFβ-stimulated fibrogenic gene expression (fig. S7H).
DISCUSSION
Our results are clinically relevant. First and foremost, we established GLRX as a promising therapeutic target for liver fibrosis. Delivery of the GLRX gene to activated HSCs using the AEAA-conjugated nanoparticles was sufficient to reverse the progression of established liver fibrosis. Most excitingly, we showed that GLRX is likely the elusive therapeutic target of the anti-fibrotic drug PFD. PFD inhibits HSC activation and liver fibrosis by targeting GLRX. PFD induces the expression of Glrx in HSCs in a Stat5-dependent manner. However, the mechanism by which PFD induces the expression and activity of Stat5 remains to be defined.
Disruption of hepatic redox homeostasis is common in liver fibrosis (8), and antioxidant supplementation has been considered for the treatment of liver fibrosis (1, 29). However, therapeutic benefits of antioxidants are yet to be observed in clinical trials. A lack of understanding of the mechanism by which oxidative stress facilitates HSC activation and liver fibrosis may have hindered the therapeutic benefit of antioxidants. Targeting cysteine oxidation by GLRX is distinct from previous studies that were mostly focused on nonspecific antioxidants. The protein residue of cysteine is required to maintain cellular redox homeostasis (12, 30). As the most susceptible protein constituent to oxidative modification (31), oxidized cysteines undergo reversible redox-based PSSG, which changes the structure and function of target proteins to protect them from further irreversible protein oxidation (32). GLRX regulates the oxidative state of protein cysteines by reversing PSSG through the liberation of GSH (33). Our study showed accumulation of PSSG accompanied by decreased expression of GLRX in both mouse and human liver fibrosis. The down-regulation of GLRX may have resulted from dysregulation of mitochondrial production of reactive oxygen species (34), but future studies are necessary to fully understand the fibrosis-responsive GLRX depletion. The inverse correlation between GLRX and fibrogenic gene expression points to an important role of protein glutathionylation in liver fibrosis. We propose that PSSG is a biomarker for liver fibrosis.
Our results strongly suggested that it is the GLRX in HSCs that plays an important role in liver fibrosis, because GLRX expression is the highest in quiescent HSCs, and the fibrosis-responsive suppression of GLRX occurred exclusively in HSCs. However, in the whole-body Glrx−/− mice, we cannot exclude the possibility that the loss of Glrx in HEPs may have also contributed to the phenotypic exhibition. We found that Glrx-depleted primary HEPs appeared to be less sensitive to CCl4- or TAA-induced toxicity at least in vitro, suggesting that the loss of Glrx in the HEPs of Glrx−/− mice may not account for the increased sensitivity to fibrosis.
Our study is mechanistic in nature. We identified Smad3 as a bona fide substrate and key effector for the anti-fibrotic activity of GLRX. GLRX interacts with Smad3, leading to sequestration of Smad3 from TGFBRI, and inhibition of Smad3 phosphorylation and nuclear translocation. Moreover, we have provided evidence that glutathionylation of Smad3 is required for its fibrogenic activity. Collectively, our data suggested that GLRX exerts its anti-fibrotic activity through resolving Smad3 oxidation. However, we cannot exclude the possibility of other GLRX substrate proteins that may have also contributed to the phenotypic exhibition. Our results suggest that there are substrate specificities in protein glutathionylation and deglutathionylation, but the mechanism of which remains to be understood.
Although PFD was effective in preventing liver fibrosis and reversing preestablished liver fibrosis in mice, its ultimate use in treating human liver fibrosis remains to be seen. It has been recognized that advanced fibrosis in human is less reversible than in rodents, likely due to the densely crosslinked and accumulated ECM over decades in patients. Current phase 2 trials are focusing on the utilization of PFD in patients with advanced liver fibrosis (ClinicalTrials.gov Identifier: NCT04099407). On the basis of the on-line information, the primary and secondary outcome measures showed at least 30% reduction of fibrosis score and improvement in serum concentrations of TGFβ1. Results from our preclinical models strongly suggest that clinical trials of PFD may have to be extended to earlier phases of liver fibrosis when HSCs have a better chance to be targeted by this drug.
Future studies are necessary to determine whether GLRX is also required for the anti-lung fibrosis effect of PFD. Activation and transdifferentiation of HSCs to myofibroblast-like cells are the key event in the progression of liver fibrosis, in which myofibroblast precursor cells are almost exclusively derived from quiescent HSCs. In contrast, recruited fibrocytes from peripheral blood are believed to be predominant in lung fibrosis (2). Administration of recombinant Glrx protein has been reported to protect mice from experimental lung fibrosis but without defined target cell types and proteins (10).
In summary, we showed that the PSSG-GLRX-Smad3 redox axis plays an essential role in HSC activation and liver fibrosis. Our results established GLRX/PSSG as both a biomarker and a therapeutic target of liver fibrosis. In addition, we have provided proof-of-concept evidence that GLRX induction by PFD or other pharmacological agents that reduces protein glutathionylation is a promising strategy to prevent and treat liver fibrosis.
MATERIALS AND METHODS
Study design
The goals of this study were to evaluate the effect of the anti-lung fibrosis drug PFD on HSC activation and liver fibrosis, to identify the therapeutic target of PFD, and to determine the mechanism by which PFD inhibits HSC activation and liver fibrosis. We found that PFD inhibited HSC activation and experimental liver fibrosis in a GLRX-dependent manner. This led to our hypothesis that the anti-fibrosis effect of PFD is mediated by GLRX, and GLRX is a novel therapeutic target of liver fibrosis. The effect of GLRX on HSC activation in vitro was determined in culture-activated or TGFβ-stimulated primary mouse and human HSCs infected with Ad-GLRX or Ad-shGlrx, or HSCs isolated from WT or Glrx−/− mice. For in vivo studies, 8-week-old male WT and Glrx−/− mice, and WT mice that bear HSC-specific GLRX overexpression or knockdown, were randomly assigned to different groups and subjected to mouse models of liver fibrosis. Molecular biology techniques were used to characterize Smad3 as a novel substrate for GLRX-mediated deglutathionylation. Patient specimens involved were collected under the shown criteria, listed in Materials and Methods. The exact sample sizes for each experimental group are listed in the respective figure legends. Sample size was estimated on the basis of sample availability, previous studies, and literatures. The investigators involved in this study were not completely blinded during treatments and liver sample collections. However, biochemical and liver fibrotic histological analyses were performed blindly. All attempts at replication were successful. No data were excluded during this study.
Human liver biopsies
Deidentified normal and cirrhotic human liver sections were purchased from Sekisui Xeno Tech (Kansas City, KS). These are formalin-fixed and paraffin-embedded liver sections from Caucasian male donors with liver fibrosis but no diabetes.
Mice and mouse models of liver fibrosis
All experiments used male mice in the C57BL/6J background. Glrx−/− mice were created using the CRISPR-Cas9 technology by Shanghai Model Organisms Centers (Shanghai, China). Mouse models of liver fibrosis were established as we have previously described (4). In the BDL model, mice were subjected to common BDL or the sham surgery for 2 weeks. In the CCl4 model, mice were injected with corn oil or CCl4 (0.5 μl/g body weight) intraperitoneally twice a week for 8 weeks. In the TAA model, mice were intraperitoneally injected with phosphate-buffered saline (PBS) or TAA (100 mg/kg body weight) three times a week for 8 weeks. The use of mice was in accordance with the University of Pittsburgh Institutional Animal Care and Use Committee.
Primary human and mouse liver cells
Normal human HEPs, HSCs, and LSECs were obtained through the Liver Tissue Cell Distribution System, Pittsburgh, Pennsylvania, which was funded by National Institutes of Health (NIH) contract #HHSN276201200017C. Primary HEPs were cultured in HepatoZYME-SFM after attachment in William E medium for 2 hours. Primary HSCs were isolated as previously reported (35) and cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS). The purity of HSCs was confirmed by vitamin A autofluorescence (4). In Fig. 4, primary mouse or human HSCs were isolated and subjected to culture-activated model by culturing them on plastic culture plates for up to 10 days. In Fig. 5, primary mouse or human HSCs were infected with Ad-GLRX or Ad-Ctrl 1 day after isolation and culture-activated for four additional days. Primary LSECs were isolated as described (36–38) and cultured in RPMI containing 10% FBS. Primary mouse HEPs, HSCs, KCs, and LSECs were isolated similarly following in situ two-step collagenase perfusion of the liver.
Histology
Liver tissues were fixed in 10% formalin overnight. Three-micrometer liver paraffin sections were prepared, then deparaffinized, and rehydrated before staining. For immunostaining of GLRX or α-SMA, antigen retrieval was performed by boiling the sections in citric acid solution for 20 min. The primary antibodies used were anti-GLRX (1:200, ab45953, Abcam) and anti–α-SMA (1:200; ab32575, Abcam). The VECTASTAIN ABC Kit (PK-6101) was used, and the staining was visualized using the DAB Peroxidase Substrate Kit (SK-4105) from Vector Laboratories (Burlingame, CA). For Sirius Red staining, hydrated liver sections were incubated in 0.1% PicroSirius Red solution for 1 hour followed by washing in 1% acetic acid solution. Immunostaining and Sirius Red staining were quantified by threshold analysis using the NIH ImageJ software.
In situ PSSG visualization and assessment of PSSG in tissue or cell extracts
PSSG was visualized in situ in liver sections or primary HSCs with the S-Glutathionylated Protein Detection Kit (catalog no. 10010721) from Cayman Chemical (Ann Arbor, MI). Briefly, hydrated tissue sections or fixed HSCs were incubated with freshly prepared blocking agent to block existing protein-free thiols. PSSG sites were enzymatically reduced to reconstitute new free thiols by PSSG reduction reagent. The reconstituted free thiols were then labeled with thiol-specific biotin-maleimide using a labeling reagent. PSSG detection fluorescein isothiocyanate (FITC) reagent was used for subsequent in situ PSSG visualization by avidin conjugation. PSSG levels in liver tissue or HSC extracts were detected using an enzymatic recycling assay as previously reported (39). Briefly, protein samples were prepared by homogenization, and proteins were precipitated by 0.6% sulfosalicylic acid. GSH was removed from PSSG by 1% NaBH4 and then neutralized with 30% metaphosphoric acid. After centrifugation, GSH released from the conjugated proteins was determined by DTNB-GSSG reductase recycling method using the EnzyChrom Glutathione Assay Kit (EGTT-100) from BioAssay Systems (Hayward, CA).
Immunofluorescence
HSCs were seeded and drug-treated in slide chambers. After fixing in 4% paraformaldehyde, cells were permeabilized in 0.3% Triton X-100 for 30 min. After being blocked in 5% goat serum for 1 hour, slides were incubated with primary antibodies against GLRX (1:200; ab45953, Abcam), α-SMA (1:200; A2547, Sigma-Aldrich), Ki67 (1:200; ab15580, Abcam), or Smad3 (1:200; ab28379, Abcam) overnight at 4°C. The slides were then washed and incubated with the secondary antibodies (ab150077, ab97035, Abcam) for 2 hours at 37°C before being mounted and analyzed using a fluorescence microscope (BZ-X810) from Keyence (Itasca, IL).
Quantitative real-time PCR
Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and reverse-transcribed with the High-Capacity cDNA Reverse Transcription Kit (No. 4368814) from Applied Biosystems/Thermo Fisher Scientific (Waltham, MA). SYBR Green–based qPCR was performed using the QuantStudio 6 Flex Real-Time PCR System. The sequences of qPCR primers are listed in table S1.
Adenovirus, plasmid construction, and cell transfection
Adenovirus-overexpressing human GLRX and the control virus have been described previously (40). Adenoviruses expressing the mutant human GLRX C23S and mouse Glrx shRNA were purchased from Vector Biolabs (Malvern, PA). The pcDNA3.1+-Stat5a plasmid was purchased from GenScript (catalog no. OMu22532D). The mouse Glrx promotor (nucleotides −437 to +83) luciferase reporter gene pGL3-Glrx, pCDH-Stat5b, pCMX-HA-human Smad2, HA-human Smad3 (FL, MH1-L, or L-MH2), and pCDH-human GLRX were cloned using standard molecular cloning techniques. Mutations of pGL3-Glrx and GLRX from cysteine to serine and Smad3 from cysteines to alanines were performed using the QuikChange Lightning Site-Directed Mutagenesis Kit (catalog no. 210519) from Agilent (Santa Clara, CA). All cloned sequences were verified by DNA sequencing. Transfection was performed using Lipofectamine 3000 from Invitrogen (Waltham, MA).
Co-IP and Western blot analysis
Protein extracts for immunoprecipitation were prepared using nondenaturing immunoprecipitation buffer supplemented with protease and phosphatase inhibitor cocktails (78440, Thermo Fisher Scientific) and 20 mM N-ethylmaleimide (E3876, Sigma-Aldrich). Following protein quantifications, primary antibodies against His-Tag (12698, Cell Signaling Technology), Smad3 (sc-101154, Santa Cruz Biotechnology), or GSH (101-A, Virogen) were added to immunoprecipitate indicated proteins using Protein A Magnetic Beads (S1425S, New England Biolabs). The beads were washed, and protein elutes were analyzed by Western blotting. For Western blotting, cells or homogenized tissues were lysed in radioimmunoprecipitation assay (RIPA) buffer. Primary antibodies for Western blotting include GLRX (ab187507), Smad3 (ab28379), and TGFBR1 (ab235178) from Abcam; α-SMA (A2547) from Sigma-Aldrich; Timp-1 (sc-21734) and Col1a1 (sc-293182) from Santa Cruz Biotechnology; HA-Tag (3724), His-Tag (12698), and p-Smad3 (9520) from Cell Signaling Technology; and ZFYVE9 (PA5-67946) and TGFBR2 (PA5-36115) from Invitrogen. Following incubation with secondary antibodies, Pierce ECL Substrate was used for signal detection.
HSC Matrigel invasion assay
Matrigel invasion of HSCs was assessed by using Corning Matrigel Basement Membrane Matrix (356234) and Corning Costar transwell cell culture inserts (CLS3422). HSCs were plated onto the upper chamber using serum-free medium. HSC invasion to the lower chamber was stimulated by medium of lower chamber containing 10% FBS and TGFβ1 (4 ng/ml) for 5 days. After fixing in 4% paraformaldehyde, cells were stained with 0.4% crystal violet solution to assess cell invasion with a microscope.
RNA-seq analysis
Total RNA was extracted with the RNeasy Mini Kit and subjected to library preparation. RNA-seq was performed by the Health Sciences Sequencing Core at the Children’s Hospital of Pittsburgh. RNA-seq analysis was performed as we have previously described (41).
ChIP assay
ChIP assay was performed as previously reported (42). Primary HSCs were cross-linked by 1.42% formaldehyde followed by quenching with 125 mM glycine. Cells were lysed by ChIP buffer with protease inhibitor cocktail. After sonication, sheared chromatin supernatants were added with normal rabbit immunoglobulin G (IgG) (2729, Cell Signaling Technology), anti-Stat5 antibody (94205, Cell Signaling Technology), or anti-Smad3 antibody (ab28379, Abcam) followed by precipitation with Protein A Magnetic Beads (S1425S, New England Biolabs). The DNA was eluted from the beads and heated to reverse cross-linking before being subjected to real-time PCR using the following ChIP primers (43): Acta2: 5′-CAAGTCCTCAGCTAATGGCC-3′ (forward) and 5′-GGGGATAAACATCCTAAGCC-3′ (reverse); Col1a1: 5′-CCTCTGCCTCTTCTTGAGAGC-3′ (forward) and 5′-GGAGAGGAGCTAAGTGTGAAGC-3′ (reverse); Glrx promoter: 5′-GTACCCACCTTACAGGGCAA-3′ (forward) and 5′-TGCATAGTGATTGGGCCTTG-3′ (reverse); Bcl2 promoter: 5′-TTGCCGAGAAGAAGGGAGAA-3′ (forward) and 5′-CGGCGGCAGATGAATTACAA-3′ (reverse); Il10 promoter: 5′-ATTGTAAAACAGGGCCATGG-3′ (forward) and 5′-GGCAGTTGGTCAGAGGAGAG-3′ (reverse); Cdkal1 promoter: 5′-GGCAAATGGATCAGTGCTCA-3′ (forward) and 5′-TCCAAACTGCGAGAACAAGC-3′ (reverse).
Promoter luciferase reporter gene assay
293T cells were cotransfected with the pGL3-Glrx or pGL3-mutant Glrx, and pcDNA3.1+-Stat5a or pCDH-Stat5b for 24 hours. LX2 and 293T cells were transfected with the Smad3-responsive luciferase reporter SBE-Luc (a gift from X. Cao, Johns Hopkins School of Medicine), together with Smad3- and/or GLRX-expressing plasmids for 24 hours in the absence or presence of TGFβ1 (5 ng/ml) (T7039, Sigma-Aldrich) stimulation. Luciferase activity was measured and normalized for transfection efficiency against β-galactosidase activity from the cotransfected pCMX-β-gal (44).
AEAA-conjugated nanoparticle preparation and characterization
AEAA-conjugated nanocarriers loaded with pCDH vector, GLRX WT plasmid, GLRX C23S plasmid, pLKO.1-puro shCon, pLKO.1-puro shGlrx (SHCLNG, Sigma-Aldrich), siCon, or siGlrx (AM16706, Ambion) were formulated by a stepwise self-assembly process as we previously reported (22). The size of nanoparticles was determined with Malvern ZetaSizer (ZEN3600, Westborough, MA).
RNA in situ hybridization
Paraffin sections were prepared using RNAscope Pretreatment Reagents (322000, 322330) and then hybridized with Mm-Col1a1 probe (319371, ACD) and Hs-GLRX probe (494451, ACD). The RNAscope Duplex Detection Kit (PN 322500) was used to visualize two target genes by two colored chromogen precipitates.
ALT release assay
After drug treatment for 24 hours, the medium of primary HEPs was collected, and the cell lysates were prepared using 1% Triton X-100. Measurement of ALT activities in the medium and cell lysates was performed using a commercial ALT assay kit (191841, Stanbio).
Statistical analysis
Prism GraphPad 7.0 software (San Diego, CA) was used. Statistical significance was evaluated by the unpaired two-tailed Student’s t test between two groups. Comparison for multiple groups was performed by using one-way analysis of variance (ANOVA) followed by Tukey’s test. Pearson correlation coefficient analysis and linear regression analysis were used to evaluate the correlation of GLRX and fibrogenic gene expression. The statistical significance was set as *P < 0.05 or **P < 0.01.
Acknowledgments
Funding: This work was supported, in part, by NIH grant ES030429 (to W.X.). W.X. was also supported in part by the Joseph Koslow Endowed Professorshsip provided by the University of Pittsburgh School of Pharmacy. Y.X. is a visiting student from Sun Yat-sen University supported by a Visiting Student Scholarship from the Government of China’s China Scholarship Council (file no. 201806380128). Author contributions: W.X. and Z.H. conceived and mentored this research. Y.X. and W.X. designed the study. Y.X., Y.L., P.X., Sihan Li, and Z.L. performed the experiments and analyzed the data. H.-c.T., X.C., J.W., H.H., M.W., M.X., S.R., Song Li, M.Z., Y.L., L.H., D.Y., and J.H. gave technical support and conceptual advice. W.X. and Y.X. wrote the manuscript. All authors edited the manuscript and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
REFERENCES AND NOTES
- 1.Bataller R., Brenner D. A., Liver fibrosis. J. Clin. Invest. 115, 209–218 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellicoro A., Ramachandran P., Iredale J. P., Fallowfield J. A., Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 14, 181–194 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Chedid M. F., Nonalcoholic steatohepatitis: The second leading indication for liver transplantation in the USA. Dig. Dis. Sci. 62, 2621–2622 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Yan J., Tung H.-C., Li S., Niu Y., Garbacz W. G., Lu P., Bi Y., Li Y., He J., Xu M., Ren S., Monga S. P., Schwabe R. F., Yang D., Xie W., Aryl hydrocarbon receptor signaling prevents activation of hepatic stellate cells and liver fibrogenesis in mice. Gastroenterology 157, 793–806.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchida T., Friedman S. L., Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 14, 397–411 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Massague J., Seoane J., Wotton D., Smad transcription factors. Genes Dev. 19, 2783–2810 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Novo E., Busletta C., Bonzo L. V., Povero D., Paternostro C., Mareschi K., Ferrero I., David E., Bertolani C., Caligiuri A., Cannito S., Tamagno E., Compagnone A., Colombatto S., Marra F., Fagioli F., Pinzani M., Parola M., Intracellular reactive oxygen species are required for directional migration of resident and bone marrow-derived hepatic pro-fibrogenic cells. J. Hepatol. 54, 964–974 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Parola M., Robino G., Oxidative stress-related molecules and liver fibrosis. J. Hepatol. 35, 297–306 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Novo E., Parola M., The role of redox mechanisms in hepatic chronic wound healing and fibrogenesis. Fibrogenesis Tissue Repair 5, S4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anathy V., Lahue K. G., Chapman D. G., Chia S. B., Casey D. T., Aboushousha R., van der Velden J. L. J., Elko E., Hoffman S. M., McMillan D. H., Jones J. T., Nolin J. D., Abdalla S., Schneider R., Seward D. J., Roberson E. C., Liptak M. D., Cousins M. E., Butnor K. J., Taatjes D. J., Budd R. C., Irvin C. G., Ho Y. S., Hakem R., Brown K. K., Matsui R., Bachschmid M. M., Gomez J. L., Kaminski N., van der Vliet A., Janssen-Heininger Y. M. W., Reducing protein oxidation reverses lung fibrosis. Nat. Med. 24, 1128–1135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janssen-Heininger Y. M., Aesif S. W., van der Velden J., Guala A. S., Reiss J. N., Roberson E. C., Budd R. C., Reynaert N. L., Anathy V., Regulation of apoptosis through cysteine oxidation: Implications for fibrotic lung disease. Ann. N. Y. Acad. Sci. 1203, 23–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsui R., Ferran B., Oh A., Croteau D., Shao D., Han J., Pimentel D. R., Bachschmid M. M., Redox regulation via glutaredoxin-1 and protein S-glutathionylation. Antioxid. Redox Signal. 32, 677–700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sgalla G., Cocconcelli E., Tonelli R., Richeldi L., Novel drug targets for idiopathic pulmonary fibrosis. Expert Rev. Respir. Med. 10, 393–405 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Philips C. A., Padsalgi G., Ahamed R., Paramaguru R., Rajesh S., George T., Mahadevan P., Augustine P., Repurposing pirfenidone for nonalcoholic steatohepatitis-related cirrhosis: A case series. J. Clin. Transl. Hepatol. 8, 100–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tada S., Nakamuta M., Enjoji M., Sugimoto R., Iwamoto H., Kato M., Nakashima Y., Nawata H., Pirfenidone inhibits dimethylnitrosamine-induced hepatic fibrosis in rats. Clin. Exp. Pharmacol. Physiol. 28, 522–527 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Di Sario A., Bendia E., Svegliati Baroni G., Ridolfi F., Casini A., Ceni E., Saccomanno S., Marzioni M., Trozzi L., Sterpetti P., Taffetani S., Benedetti A., Effect of pirfenidone on rat hepatic stellate cell proliferation and collagen production. J. Hepatol. 37, 584–591 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Salazar-Montes A., Ruiz-Corro L., Lopez-Reyes A., Castrejon-Gomez E., Armendariz-Borunda J., Potent antioxidant role of pirfenidone in experimental cirrhosis. Eur. J. Pharmacol. 595, 69–77 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Zhao X. Y., Zeng X., Li X. M., Wang T. L., Wang B. E., Pirfenidone inhibits carbon tetrachloride- and albumin complex-induced liver fibrosis in rodents by preventing activation of hepatic stellate cells. Clin. Exp. Pharmacol. Physiol. 36, 963–968 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Komiya C., Tanaka M., Tsuchiya K., Shimazu N., Mori K., Furuke S., Miyachi Y., Shiba K., Yamaguchi S., Ikeda K., Ochi K., Nakabayashi K., Hata K. I., Itoh M., Suganami T., Ogawa Y., Antifibrotic effect of pirfenidone in a mouse model of human nonalcoholic steatohepatitis. Sci. Rep. 7, 44754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu B. M., Kang K., Yu J. H., Chen W., Smith H. E., Lee D., Sun H. W., Wei L., Hennighausen L., Genome-wide analyses reveal the extent of opportunistic STAT5 binding that does not yield transcriptional activation of neighboring genes. Nucleic Acids Res. 40, 4461–4472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller J., Sperl B., Reindl W., Kiessling A., Berg T., Discovery of chromone-based inhibitors of the transcription factor STAT5. Chembiochem 9, 723–727 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Hu M., Zhou X., Wang Y., Guan K., Huang L., Relaxin-FOLFOX-IL-12 triple combination therapy engages memory response and achieves long-term survival in colorectal cancer liver metastasis. J. Control. Release 319, 213–221 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Song W. T., Hu M. Y., An S., Xu L. G., Li J. J., Kinghorn K. A., Liu R. H., Huang L., Nanoparticle-mediated HMGA1 silencing promotes lymphocyte infiltration and boosts checkpoint blockade immunotherapy for cancer. Adv. Funct. Mater. 28, 1802847 (2018). [Google Scholar]
- 24.Hu M., Wang Y., Xu L., An S., Tang Y., Zhou X., Li J., Liu R., Huang L., Relaxin gene delivery mitigates liver metastasis and synergizes with check point therapy. Nat. Commun. 10, 2993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D., Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 10, 1941–1988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukazaki T., Chiang T. A., Davison A. F., Attisano L., Wrana J. L., SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell 95, 779–791 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Derynck R., Zhang Y. E., Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425, 577–584 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Macias-Barragan J., Sandoval-Rodriguez A., Navarro-Partida J., Armendariz-Borunda J., The multifaceted role of pirfenidone and its novel targets. Fibrogenesis Tissue Repair 3, 16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoyama T., Paik Y. H., Watanabe S., Laleu B., Gaggini F., Fioraso-Cartier L., Molango S., Heitz F., Merlot C., Szyndralewiez C., Page P., Brenner D. A., Nicotinamide adenine dinucleotide phosphate oxidase in experimental liver fibrosis: GKT137831 as a novel potential therapeutic agent. Hepatology 56, 2316–2327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J., Berisa M., Schwörer S., Qin W., Cross J. R., Thompson C. B., Transsulfuration activity can support cell growth upon extracellular cysteine limitation. Cell Metab. 30, 865–876.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forman H. J., Fukuto J. M., Torres M., Redox signaling: Thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am. J. Physiol. Cell Physiol. 287, C246–C256 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Kuipers I., Guala A. S., Aesif S. W., Konings G., Bouwman F. G., Mariman E. C., Wouters E. F., Janssen-Heininger Y. M., Reynaert N. L., Cigarette smoke targets glutaredoxin 1, increasing s-glutathionylation and epithelial cell death. Am. J. Respir. Cell Mol. Biol. 45, 931–937 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen E. M., Mieyal J. J., Protein-thiol oxidation and cell death: Regulatory role of glutaredoxins. Antioxid. Redox Signal. 17, 1748–1763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diamond D. L., Jacobs J. M., Paeper B., Proll S. C., Gritsenko M. A., Carithers R. L. Jr., Larson A. M., Yeh M. M., Camp D. G. II, Smith R. D., Katze M. G., Proteomic profiling of human liver biopsies: Hepatitis C virus–induced fibrosis and mitochondrial dysfunction. Hepatology 46, 649–657 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Mederacke I., Dapito D. H., Affo S., Uchinami H., Schwabe R. F., High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aparicio-Vergara M., Tencerova M., Morgantini C., Barreby E., Aouadi M., Isolation of Kupffer cells and hepatocytes from a single mouse liver. Methods Mol. Biol. 1639, 161–171 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Liu J., Huang X., Werner M., Broering R., Yang D., Lu M., Advanced method for isolation of mouse hepatocytes, liver sinusoidal endothelial cells, and Kupffer cells. Methods Mol. Biol. 1540, 249–258 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Cabral F., Miller C. M., Kudrna K. M., Hass B. E., Daubendiek J. G., Kellar B. M., Harris E. N., Purification of hepatocytes and sinusoidal endothelial cells from mouse liver perfusion. J. Vis. Exp. 2018, 56993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahman I., Kode A., Biswas S. K., Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Song J. J., Lee Y. J., Differential role of glutaredoxin and thioredoxin in metabolic oxidative stress-induced activation of apoptosis signal-regulating kinase 1. Biochem. J. 373, 845–853 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Yang B., Zhang M., Guo W., Wu Z., Wang Y., Jia L., Li S.; N. Cancer Genome Atlas Research, Xie W., Yang D., lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that Interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 33, 706–720.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson J. D., Denisenko O., Bomsztyk K., Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Duran A., Hernandez E. D., Reina-Campos M., Castilla E. A., Subramaniam S., Raghunandan S., Roberts L. R., Kisseleva T., Karin M., Diaz-Meco M. T., Moscat J., p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell 30, 595–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Y., Xie Y., Xu M., Li L., Selcer K. W., Oberly P. J., Poloyac S. M., Wang H., Li C., Dong F., Yu C., Xie W., Hepatic steroid sulfatase critically determines estrogenic activities of conjugated equine estrogens in human cells in vitro and in mice. J. Biol. Chem. 294, 12112–12121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1