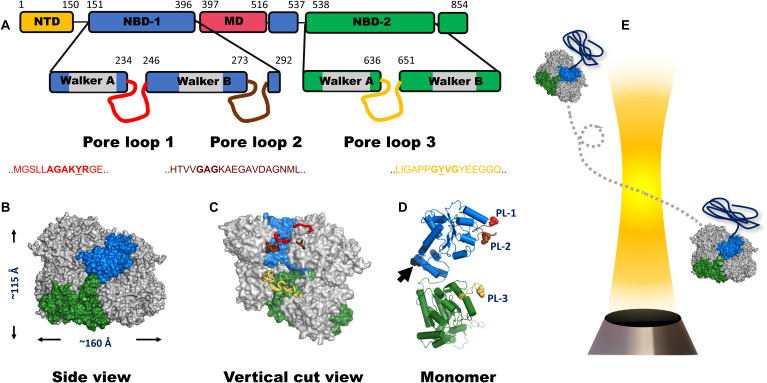
Fig. 1. ClpB domain organization and structure.
(A) Domain organization of the monomer of ClpB. NTD, N-terminal domain; NBD, nucleotide-binding domain; MD, middle domain. The location and sequence of pore loops are indicated on each NBD. The numbers mark residue positions in the sequence. A ClpB variant that lacks the NTD is used in this study. This variant actually occurs naturally and is fully functional. (B) Side view of the hexameric model of E. coli ClpB reconstructed from cryo-EM data (PDB: 6OAX). One protomer (protomer A) is highlighted using the same color code as in (A). The MD and the NTD are not resolved in this structure; thus, they are not shown. (C) Vertical cut view of the hexameric structure in (B). PL1, PL2, and PL3 of one protomer are colored in red, brown, and yellow, respectively. PL1 and PL2 are not fully resolved. (D) Monomeric structure of E. coli ClpB (PDB: 6OAX, protomer A) with α-helices represented as cylinders and pore loops shown in the same color as in (C). The spheres on pore loops represent the conserved tyrosine residues of PL1 (red) and PL3 (yellow) and the alanine residue of the motif GAG in PL2 (brown). Black arrow points to position P368, which is equivalent to position S359 in T. thermophilus ClpB, used in our smFRET experiments. (E) Freely diffusing molecules of ClpB, assembled with a single double-labeled protomer per hexamer (see Methods), emit bursts of photons as they pass through a focused laser beam, from which the FRET efficiency is calculated.